Attached files
| file | filename |
|---|---|
| 8-K - 8-K - Cardiff Oncology, Inc. | a13-16871_38k.htm |
Exhibit 99.1
|
|
A better clinical pathway for cancer monitoring Antonius Schuh, Ph.D. | Chief Executive Officer July 2013 |
|
|
Forward-Looking Statements DISCLAIMER Statements in this presentation about the Company's expectations, applications of its technology, markets, launch of tests and other statements that are not historical facts are "forward-looking statements" within the meaning of Section 27A of the Securities Act of 1933 and Section 21E of the Securities Exchange Act of 1934 and are based on management's current beliefs, assumptions, estimates and projections. Actual results may differ materially from those projected in the forward-looking statements for various reasons, including risks associated with product and test development, test transfer to contracting labs, government regulation, market acceptance, limited commercial experience, dependence on key personnel, obtaining financing and other factors discussed in the Company's periodic reports filed with the Securities and Exchange Commission. |
|
|
Our goal: Towards better treatment for cancer INTRODUCTION A better clinical pathway for cancer monitoring Novel clinical utility: non-invasive, near real-time detection of oncogene mutations for any tumor type |
|
|
Welcome to Trovagene, Inc. INTRODUCTION INTRODUCTION Addresses a high unmet need in cancer by enabling easier, more frequent monitoring of emergence of oncogene mutation status, disease progression and recurrence Unique IP around cell-free DNA in urine creates a powerful core diagnostic platform for the non-invasive detection of oncogene mutations, with urine providing the competitive advantages of ease of sampling and large volume Cancer monitoring based on urine testing is now ready for clinical use as improved PCR technologies and next generation sequencing platforms are available at much reduced cost CLIA certified, CAP accredited high complexity diagnostic laboratory to offer diagnostic services Technology development collaborations with leading corporate partners NASDAQ: TROV $111M market cap* Molecular Diagnostic Specialist founded in 1999 NASDAQ listing 2012 $55M invested in R&D and IP Focused on oncology and infectious disease *Correct on 19 June, 2013 |
|
|
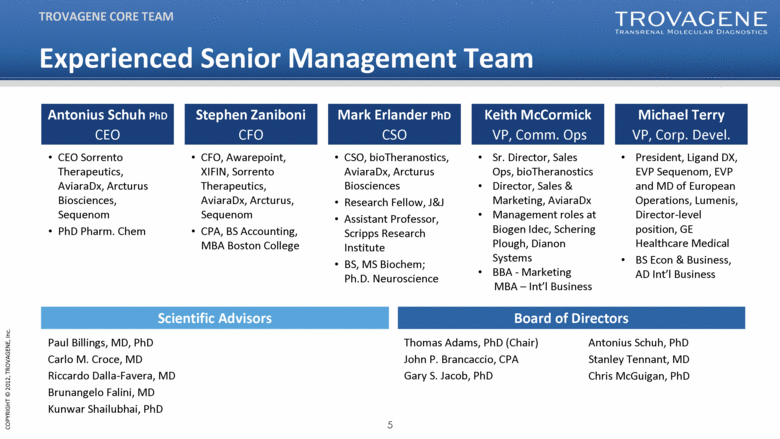
TROVAGENE CORE TEAM Antonius Schuh PhD CEO CEO Sorrento Therapeutics, AviaraDx, Arcturus Biosciences, Sequenom PhD Pharm. Chem Stephen Zaniboni CFO Mark Erlander PhD CSO Keith McCormick VP, Comm. Ops Michael Terry VP, Corp. Devel. Thomas Adams, PhD (Chair) John P. Brancaccio, CPA Gary S. Jacob, PhD CFO, Awarepoint, XIFIN, Sorrento Therapeutics, AviaraDx, Arcturus, Sequenom CPA, BS Accounting, MBA Boston College CSO, bioTheranostics, AviaraDx, Arcturus Biosciences Research Fellow, J&J Assistant Professor, Scripps Research Institute BS, MS Biochem; Ph.D. Neuroscience Sr. Director, Sales Ops, bioTheranostics Director, Sales & Marketing, AviaraDx Management roles at Biogen Idec, Schering Plough, Dianon Systems BBA - Marketing MBA – Int’l Business President, Ligand DX, EVP Sequenom, EVP and MD of European Operations, Lumenis, Director-level position, GE Healthcare Medical BS Econ & Business, AD Int’l Business Paul Billings, MD, PhD Carlo M. Croce, MD Riccardo Dalla-Favera, MD Brunangelo Falini, MD Kunwar Shailubhai, PhD Antonius Schuh, PhD Stanley Tennant, MD Chris McGuigan, PhD Scientific Advisors Board of Directors Experienced Senior Management Team |
|
|
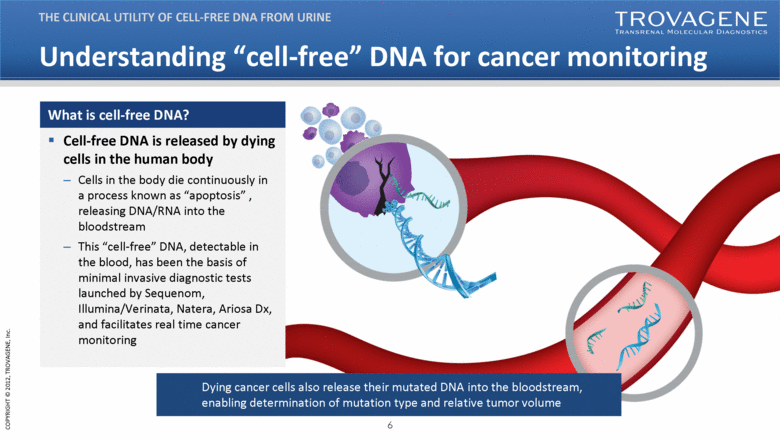
Understanding “cell-free” DNA for cancer monitoring THE CLINICAL UTILITY OF CELL-FREE DNA FROM URINE Cell-free DNA is released by dying cells in the human body Cells in the body die continuously in a process known as “apoptosis” , releasing DNA/RNA into the bloodstream This “cell-free” DNA, detectable in the blood, has been the basis of minimal invasive diagnostic tests launched by Sequenom, Illumina/Verinata, Natera, Ariosa Dx, and facilitates real time cancer monitoring What is cell-free DNA? Dying cancer cells also release their mutated DNA into the bloodstream, enabling determination of mutation type and relative tumor volume |
|
|
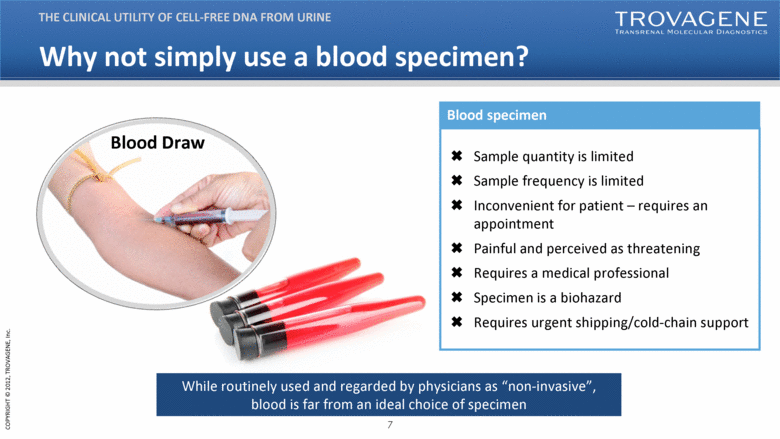
Why not simply use a blood specimen? THE CLINICAL UTILITY OF CELL-FREE DNA FROM URINE Blood Draw Sample quantity is limited Sample frequency is limited Inconvenient for patient – requires an appointment Painful and perceived as threatening Requires a medical professional Specimen is a biohazard Requires urgent shipping/cold-chain support While routinely used and regarded by physicians as “non-invasive”, blood is far from an ideal choice of specimen Blood specimen |
|
|
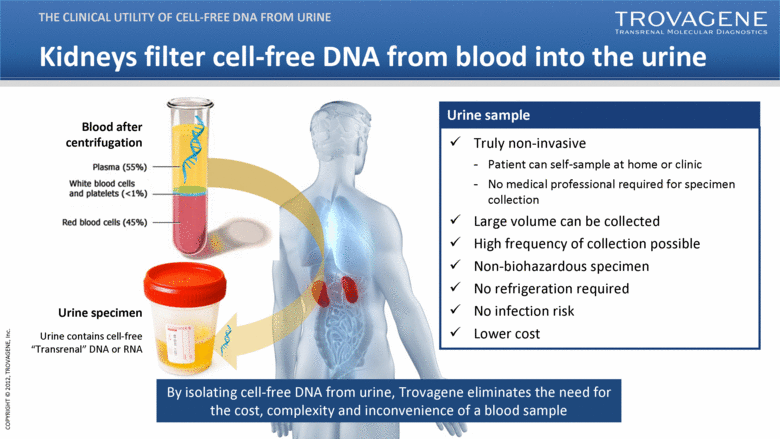
Kidneys filter cell-free DNA from blood into the urine THE CLINICAL UTILITY OF CELL-FREE DNA FROM URINE By isolating cell-free DNA from urine, Trovagene eliminates the need for the cost, complexity and inconvenience of a blood sample Truly non-invasive Patient can self-sample at home or clinic No medical professional required for specimen collection Large volume can be collected High frequency of collection possible Non-biohazardous specimen No refrigeration required No infection risk Lower cost Urine contains cell-free “Transrenal” DNA or RNA Urine sample Blood after centrifugation Urine specimen |
|
|
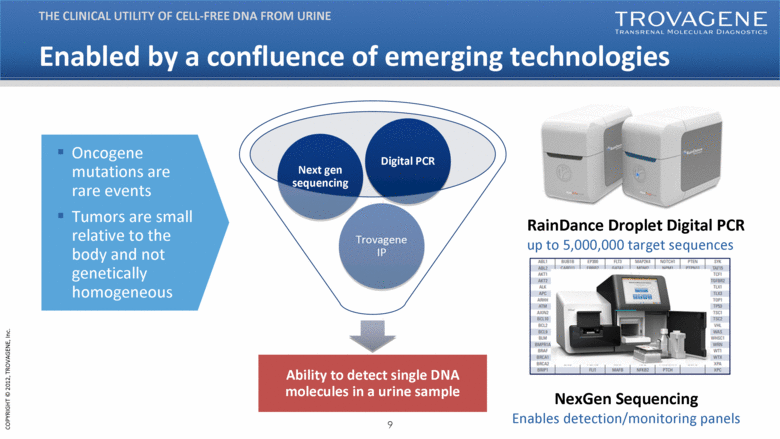
Enabled by a confluence of emerging technologies Ability to detect single DNA molecules in a urine sample Trovagene IP Next gen sequencing Digital PCR THE CLINICAL UTILITY OF CELL-FREE DNA FROM URINE RainDance Droplet Digital PCR up to 5,000,000 target sequences Oncogene mutations are rare events Tumors are small relative to the body and not genetically homogeneous NexGen Sequencing Enables detection/monitoring panels |
|
|
Why Cell-free DNA? Tumors are heterogeneous in their tumor mutational make-up Would need multiple tissue/tumor biopsies to determine heterogeneity and optimal therapy Cell-free DNA from tumors is released into the blood plasma after cell-death Previously established that cell-free DNA increases with cancer stage in plasma Technology now available to detect mutations: Digital droplet PCR and Next Generation Sequencing Platforms THE CLINICAL UTILITY OF CELL-FREE DNA FROM URINE Cell-free DNA gives a global view of tumor heterogeneity for metastatic cancer patients Detection of specific tumor mutations Monitoring of tumor load/response to therapy Detection of emerging tumor resistant mutations/new directed therapy Clinical Utility |
|
|
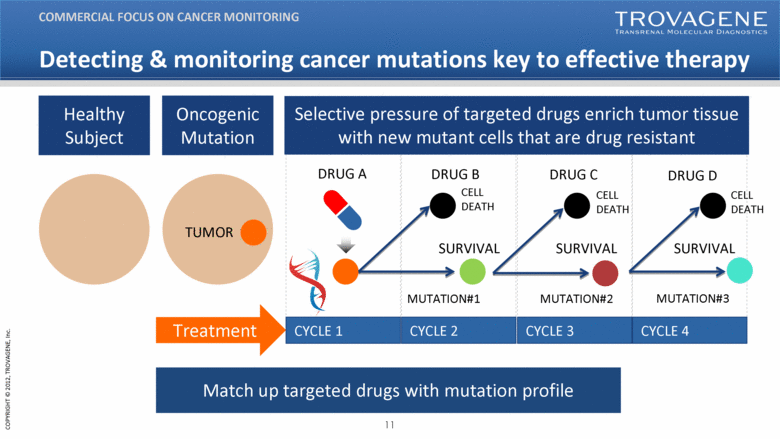
Healthy Subject Oncogenic Mutation DRUG A CYCLE 1 CYCLE 2 CYCLE 3 CYCLE 4 SURVIVAL TUMOR MUTATION#1 Selective pressure of targeted drugs enrich tumor tissue with new mutant cells that are drug resistant Match up targeted drugs with mutation profile Treatment SURVIVAL DRUG B DRUG C DRUG D SURVIVAL CELL DEATH CELL DEATH CELL DEATH MUTATION#2 MUTATION#3 Detecting & monitoring cancer mutations key to effective therapy COMMERCIAL FOCUS ON CANCER MONITORING |
|
|
Opportunity to implement monitoring of tumor dynamics, demonstrated in plasma High signal consistent with metastatic patient CLINICAL DEVELOPMENT PROGRAM Patient responds to therapy Disease progression Patients responds to therapy switch Hormonal therapy No response to hormonal RX 1Dawson et al., NEJM 2013 Online, DOI: 10.1056 2Su et al., Ann N Y Acad Sci 2008, 1137:197-206 Cell-free DNA was the only marker that predicted response CELL-FREE DNA / PIK3CA CA 15-3 ANTIGEN CIRCULATING TUMOR CELLS |
|
|
Building value through clinical validation PHASE 1 Initial studies demonstrating concordance with the existence of cancer mutations between tumor, blood and urine. Opportunity for diagnosis when a biopsy is not an option for example. Diagnostic PHASE 2 Tracking cancer mutation trends quantitatively and correlating these trends to specific treatment responses. Drug-Diagnostic PHASE 3 Requires extensive clinical validation tracking patients from cancer diagnosis through treatment Utilize oncogene mutation tracking to make specific treatment decisions Positive results demonstration improved quality and extension of life, with reduced medical costs Clinical Standard of Care $100-$200M Revenue Multi $B Revenue Improved patient outcomes $500+M Revenue QUANTITATIVE validation QUALITATIVE validation CLINICAL DEVELOPMENT PROGRAM |
|
|
Clinical Study: Detecting & Monitoring Tumor Mutations in Cell-free DNA from Urine in Metastatic Cancer Patients Title: KRAS and BRAF Mutational Analysis of Cell-free DNA from Urine in Patients with Advanced Cancers Principal Investigator: Filip Janku MD PhD; Investigational Cancer Therapeutics (Phase I Clinical Trials Program) MD Anderson Cancer Center Objective 1: To measure the degree of concordance between results of DNA mutation analysis from urine samples and tumor tissue. Objective 2: To assess treatment outcomes, such as response rate (RR), stable disease longer than 6 months (SD >6 months), progression-free survival (PFS), and overall survival (OS) with respect to mutation profile. Objective 3: To assess mutation status in DNA in patients at multiple time-points. Demonstrating utility of urine-based DNA mutational testing CLINICAL DEVELOPMENT PROGRAM |
|
|
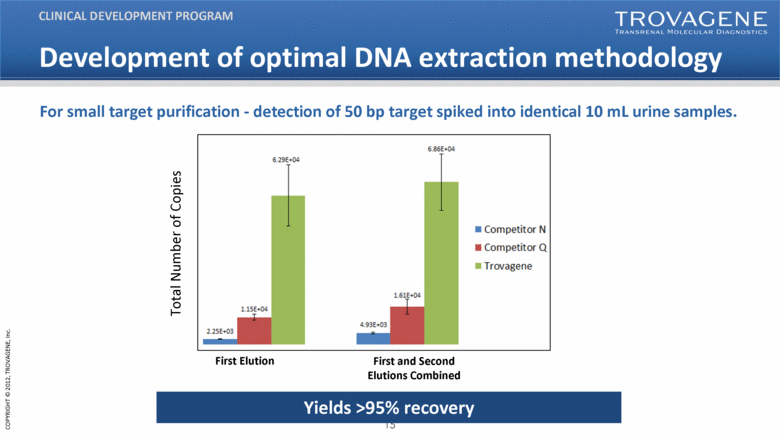
Development of optimal DNA extraction methodology CLINICAL DEVELOPMENT PROGRAM For small target purification - detection of 50 bp target spiked into identical 10 mL urine samples. First Elution First and Second Elutions Combined Total Number of Copies Yields >95% recovery |
|
|
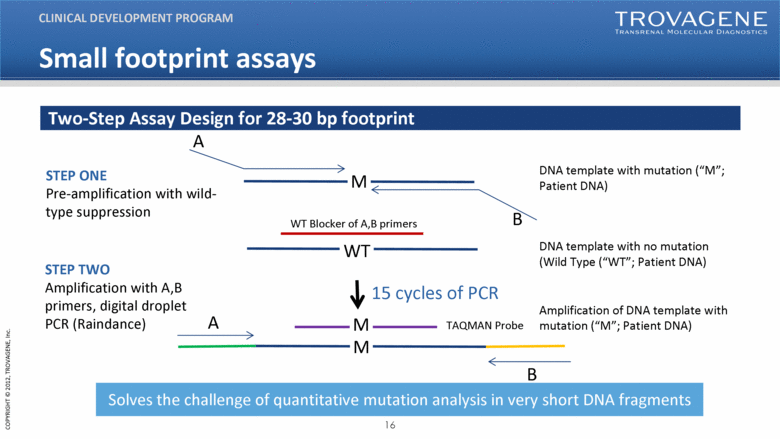
STEP ONE Pre-amplification with wild-type suppression DNA template with mutation (“M”; Patient DNA) A B STEP TWO Amplification with A,B primers, digital droplet PCR (Raindance) DNA template with no mutation (Wild Type (“WT”; Patient DNA) WT Blocker of A,B primers M WT A B M M TAQMAN Probe Amplification of DNA template with mutation (“M”; Patient DNA) 15 cycles of PCR Two-Step Assay Design for 28-30 bp footprint CLINICAL DEVELOPMENT PROGRAM Solves the challenge of quantitative mutation analysis in very short DNA fragments Small footprint assays |
|
|
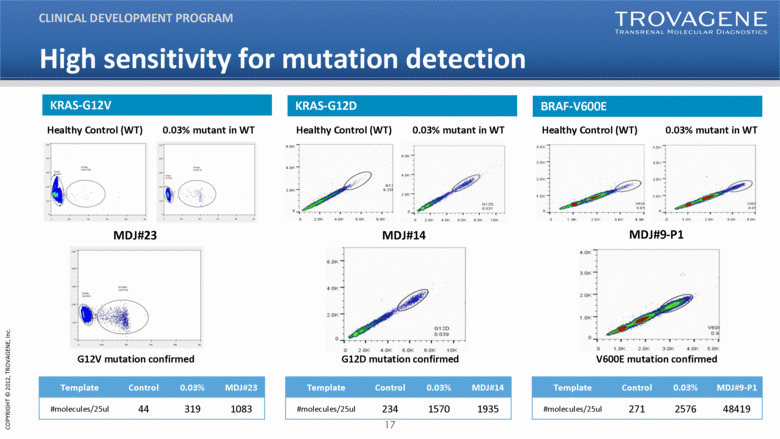
0.03% mutant in WT Healthy Control (WT) G12V mutation confirmed MDJ#23 MDJ#9-P1 V600E mutation confirmed MDJ#14 G12D mutation confirmed 0.03% mutant in WT Healthy Control (WT) 0.03% mutant in WT Healthy Control (WT) Template Control 0.03% MDJ#23 #molecules/25ul 44 319 1083 Template Control 0.03% MDJ#14 #molecules/25ul 234 1570 1935 Template Control 0.03% MDJ#9-P1 #molecules/25ul 271 2576 48419 CLINICAL DEVELOPMENT PROGRAM KRAS-G12V KRAS-G12D BRAF-V600E High sensitivity for mutation detection |
|
|
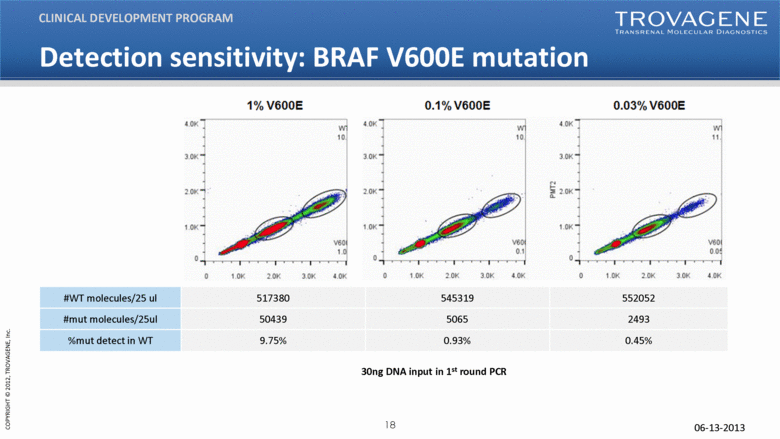
06-13-2013 30ng DNA input in 1st round PCR Detection sensitivity: BRAF V600E mutation CLINICAL DEVELOPMENT PROGRAM #WT molecules/25 ul 517380 545319 552052 #mut molecules/25ul 50439 5065 2493 %mut detect in WT 9.75% 0.93% 0.45% |
|
|
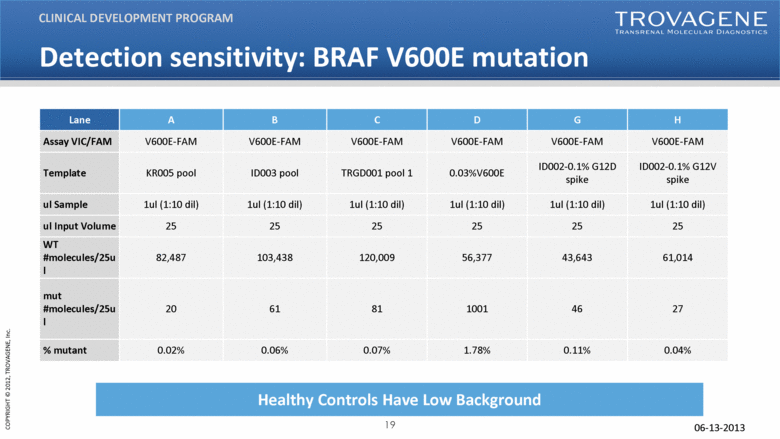
06-13-2013 30ng DNA input in 1st round PCR Detection sensitivity: BRAF V600E mutation CLINICAL DEVELOPMENT PROGRAM Healthy Controls Have Low Background Lane A B C D G H Assay VIC/FAM V600E-FAM V600E-FAM V600E-FAM V600E-FAM V600E-FAM V600E-FAM Template KR005 pool ID003 pool TRGD001 pool 1 0.03%V600E ID002-0.1% G12D spike ID002-0.1% G12V spike ul Sample 1ul (1:10 dil) 1ul (1:10 dil) 1ul (1:10 dil) 1ul (1:10 dil) 1ul (1:10 dil) 1ul (1:10 dil) ul Input Volume 25 25 25 25 25 25 WT #molecules/25ul 82,487 103,438 120,009 56,377 43,643 61,014 mut #molecules/25ul 20 61 81 1001 46 27 % mutant 0.02% 0.06% 0.07% 1.78% 0.11% 0.04% |
|
|
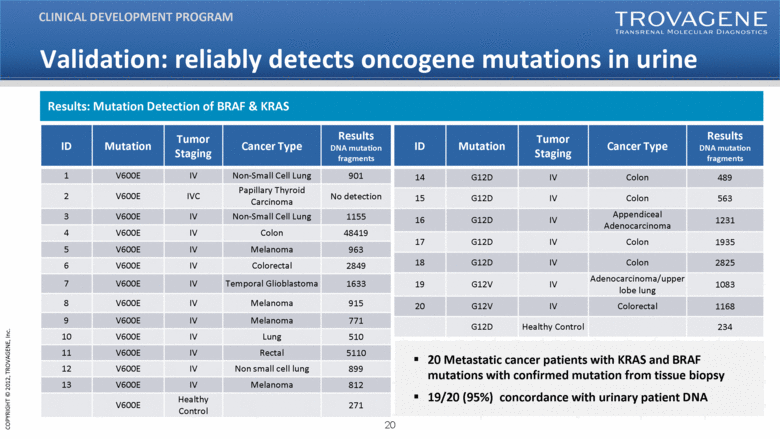
ID Mutation Tumor Staging Cancer Type Results DNA mutation fragments 14 G12D IV Colon 489 15 G12D IV Colon 563 16 G12D IV Appendiceal Adenocarcinoma 1231 17 G12D IV Colon 1935 18 G12D IV Colon 2825 19 G12V IV Adenocarcinoma/upper lobe lung 1083 20 G12V IV Colorectal 1168 G12D Healthy Control 234 20 Metastatic cancer patients with KRAS and BRAF mutations with confirmed mutation from tissue biopsy 19/20 (95%) concordance with urinary patient DNA ID Mutation Tumor Staging Cancer Type Results DNA mutation fragments 1 V600E IV Non-Small Cell Lung 901 2 V600E IVC Papillary Thyroid Carcinoma No detection 3 V600E IV Non-Small Cell Lung 1155 4 V600E IV Colon 48419 5 V600E IV Melanoma 963 6 V600E IV Colorectal 2849 7 V600E IV Temporal Glioblastoma 1633 8 V600E IV Melanoma 915 9 V600E IV Melanoma 771 10 V600E IV Lung 510 11 V600E IV Rectal 5110 12 V600E IV Non small cell lung 899 13 V600E IV Melanoma 812 V600E Healthy Control 271 Validation: reliably detects oncogene mutations in urine CLINICAL DEVELOPMENT PROGRAM Results: Mutation Detection of BRAF & KRAS |
|
|
Content Rich Ability to detect multiple mutations in a single run and any nucleotide at each position 9 KRAS mutations in a single assay: G12A, G12C, G12D, G12F, G12R, G12S, G12V, G13C, G13D Target Multiplexing Multiple Genes and multiple sites per gene can be interrogated The potential to monitor dozens of mutations in many genes at once Higher Throughput Hundreds of samples can be run simultaneously 142 samples were run in under a week for all 9 codon 12 and 13 KRAS mutations on a single sequencing run Lower Cost Multiplexing samples and targets creates a cost effective assay At $1000 per sequencing run, 142 samples can be evaluated at less than $8 per sample Maintain Sensitivity PCR enrichment techniques can provide a highly sensitive assay Rivals the sensitivity of qPCR and ddPCR since identical enrichment techniques are utilized CLINICAL DEVELOPMENT PROGRAM Next Gen Sequencing would... As proof of principle we developed a 31bp KRAS assay that can detect 9 different mutations in 142 samples in less than a week with sensitivity of less the 0.1% enable us to offer to oncologists a non-invasive, real-time detection and monitoring of ALL possible oncogene mutations in a patient |
|
|
CLINICAL DEVELOPMENT PROGRAM ...and provides generic assay design for all mutations Anatomy of a KRAS Next Gen Sequencing Assay Enables unsupervised query for any mutation, eliminating the need for the creation of individual assays for specific mutations. |
|
|
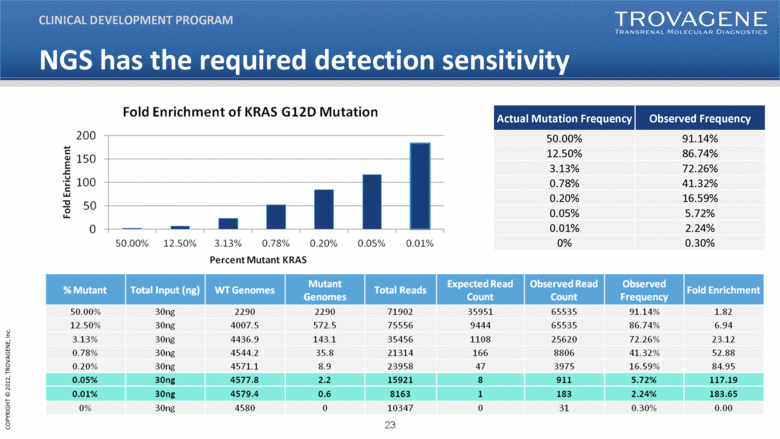
Actual Mutation Frequency Observed Frequency 50.00% 91.14% 12.50% 86.74% 3.13% 72.26% 0.78% 41.32% 0.20% 16.59% 0.05% 5.72% 0.01% 2.24% 0% 0.30% CLINICAL DEVELOPMENT PROGRAM NGS has the required detection sensitivity % Mutant Total Input (ng) WT Genomes Mutant Genomes Total Reads Expected Read Count Observed Read Count Observed Frequency Fold Enrichment 50 00% 30ng 2290 2290 71902 35951 65535 91 14% 1 82 50.00% 91.14% 1.82 12.50% 30ng 4007.5 572.5 75556 9444 65535 86.74% 6.94 3.13% 30ng 4436.9 143.1 35456 1108 25620 72.26% 23.12 0.78% 30ng 4544.2 35.8 21314 166 8806 41.32% 52.88 0.20% 30ng 4571.1 8.9 23958 47 3975 16.59% 84.95 0.05% 30ng 4577.8 2.2 15921 8 911 5.72% 117.19 0.01% 30ng 4579.4 0.6 8163 1 183 2.24% 183.65 0% 30ng 4580 0 10347 0 31 0.30% 0.00 |
|
|
53 initial targets: can be assessed using 9 designs multiplexed in a single well. The largest amplicon fragment is only 34bp. Another control set can be added for quantification purposes 1 KRAS assay 9 mutations 1 NRAS assay 7 mutations 1 BRAF 4 mutations 2 PIK3CA assay 4 mutations 2 EGFR assays 3 mutations 2 EGFR deletion assays 26 deletions EGFR T790M 1 mutation CLINICAL DEVELOPMENT PROGRAM Next steps for a first NGS-based offering We have designed ultra-sensitive short amplicon assays which have versatility for application not only in urine, but also in plasma & saliva Assay Design for version 1: Oncogene Mutation Panel Clinically Actionable Mutations |
|
|
Core IP extends into assay detection and development |
|
|
Trovagene’s Proprietary HPV Test |
|
|
HPV in Urine: market dynamics Market Opportunity Estimated 11-12M HPV tests run annually (US) 1 Represents only about 35% market penetration 8.7 – 14.2% of cases come back as positive2 Repeat HPV testing often performed at 6 and 12 months HPV 1. McEvoy and Farmer – Licensed Market Research Report – Anatomic Pathology Markets in the US – Cervical Cancer Edition – 2011 – p 305-310 2. Castle et al. Clinical Human Papilloma Virus Detection Forecasts Cervical Cancer Risk in Women Over 18 Years of Follow Up. J Clin Oncol, 30 Jul 2012 Advantages Unique primer pair amplifying the E1 region provides Freedom-To-Operate (FTO) for molecular HPV testing E1 region offers novel, proprietary approach to high risk HPV identification Simplicity of testing: only 1 PCR reaction identifies all high-risk HPV subtypes Uses Trovagene’s patented method for DNA isolation |
|
|
Validating analytical performance of our HPV assay Europe (UK) Queen Mary University of London PREDICTORS 4 Study - 500 patient cohort - samples currently being processed HPV Brazil Barretos Cancer Hospital – largest cancer hospital in Brazil 350 patient cohort - enrollment underway India Simbiosys Bioware / Metropolis 320 patient cohort (study completed) Strand Life Science Study ongoing International Validation Studies |
|
|
2012 achievements Acquired CLIA Laboratory CLIA-certified, CAP-accredited high complexity molecular diagnostic lab Strengthened Patent Portfolio Issued U.S. Patent for NPM Mutants to Diagnose and Monitor Acute Myeloid Leukemia Issued European Patent for Detection of Pathogenic Infections Improved Liquidity and Access to Capital Completed Public Offering of $10 million and began trading on NASDAQ Added to the Russell Microcap and the MSCI Microcap Index Closed Private Placement of $4.4 million Initiated KRAS Mutation Detection Development Program Commenced Validation Program and Commenced Study at MD Anderson Cancer Center for Transrenal KRAS Mutation Detection in Pancreatic Cancer Expanded of High Risk HPV Carrier Screening Development Program Partnered with Strand Life Sciences to Validate and Offer Urine-based HPV Screening Test in India Partnered with Barretos Cancer Hospital to Evaluate Urine-based HPV Assay in Brazil COMMERCIAL DEVELOPMENT UPDATE |
|
|
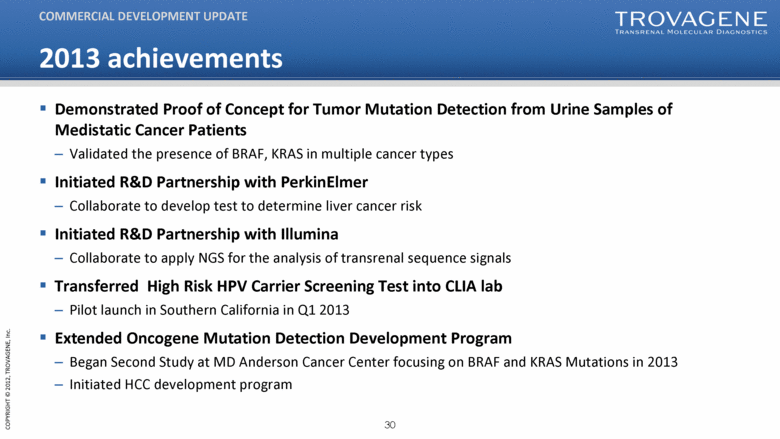
2013 achievements Demonstrated Proof of Concept for Tumor Mutation Detection from Urine Samples of Medistatic Cancer Patients Validated the presence of BRAF, KRAS in multiple cancer types Initiated R&D Partnership with PerkinElmer Collaborate to develop test to determine liver cancer risk Initiated R&D Partnership with Illumina Collaborate to apply NGS for the analysis of transrenal sequence signals Transferred High Risk HPV Carrier Screening Test into CLIA lab Pilot launch in Southern California in Q1 2013 Extended Oncogene Mutation Detection Development Program Began Second Study at MD Anderson Cancer Center focusing on BRAF and KRAS Mutations in 2013 Initiated HCC development program COMMERCIAL DEVELOPMENT UPDATE |
|
|
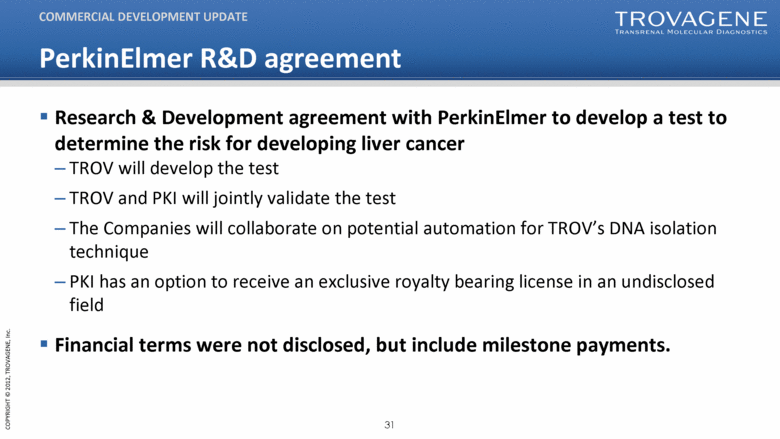
PerkinElmer R&D agreement Research & Development agreement with PerkinElmer to develop a test to determine the risk for developing liver cancer TROV will develop the test TROV and PKI will jointly validate the test The Companies will collaborate on potential automation for TROV’s DNA isolation technique PKI has an option to receive an exclusive royalty bearing license in an undisclosed field Financial terms were not disclosed, but include milestone payments. COMMERCIAL DEVELOPMENT UPDATE |
|
|
Illumina R&D agreement Goal – POC to develop library for NGS (next generation sequencing) from cell-free DNA isolated from urine Strategic imperative to establish TrNAs on NGS NGS will become a platform standard in the industry TROV to gain significant domain knowledge from market leader in NGS Illumina to provide samples and sequencing Trovagene to provide DNA extraction and purification COMMERCIAL DEVELOPMENT UPDATE Multiphase collaboration signed |
|
|
2013 milestones Expand clinical study program for qualitative validation of concordance with the existence of cancer mutations Submit abstract(s) from ongoing clinical studies Prepare draft manuscript on mutation detection for submission to peer reviewed journal Launch oncogene mutation testing in the CLIA laboratory Initiate at least two R&D partnerships with strategic partners Complete significant clinical study for urine-based HPV assay COMMERCIAL DEVELOPMENT UPDATE |
|
|
For further information Please contact: Antonius Schuh, CEO aschuh@trovagene.com Stephen Zaniboni, CFO szaniboni@trovagene.com |