Attached files
| file | filename |
|---|---|
| 8-K - 8-K - Foghorn Therapeutics Inc. | fhtx-20210714.htm |

Targeting the Chromatin Regulatory System Broadening the Impact of Precision Medicines for Oncology and Other Diseases July 2021

Disclaimer, Confidentiality, and Forward-Looking StatementsThis presentation contains forward-looking statements. All statements other than statements of historical facts contained in this presentation are forward-looking statements. In some cases, you can identify forward-looking statements by terms such as “may,” “will,” “should,” “expect,” “plan,” “anticipate,” “could,” “intend,” “target,” “project,” “contemplate,” “believe,” “estimate,” “predict,” “potential” or “continue” or the negative of these terms or other similar expressions, although not all forward-looking statements contain these words. Forward-looking statements include, but are not limited to, statements concerning: the initiation, timing, progress and results of our research and development programs and preclinical and clinical trials, our ability to advance product candidates that we may develop and successfully complete preclinical and clinical studies; our ability to leverage our initial programs to develop additional product candidates using our Gene Traffic Control Platform; the impact of the COVID-19 pandemic in our and our collaborators’ business operations, including our research and development programs and preclinical studies; developments related to our competitors and our industry; our ability to expand the target populations of our programs and the availability of patients for clinical testing; our ability to obtain regulatory approval for FHD-286, FHD-609 and any future product candidates from the FDA and other regulatory authorities; our ability to identify and enter into future license agreements and collaborations; our ability to continue to rely on our CDMOs and CROs for our manufacturing and research needs; regulatory developments in the United States and foreign countries; our ability to attract and retain key scientific and management personnel; the scope of protection we are able to establish, maintain and enforce for intellectual property rights covering FHD-286 and FHD-609, our future products and our Gene Traffic Control Platform; and our use of proceeds from our initial public offering, estimates of our expenses, capital requirements and needs for additional financing. Any forward-looking statements represent the Company’s views only as of today and should not be relied upon as representing its views as of any subsequent date. The Company explicitly disclaims any obligation to update any forward-looking statements. The Company’s business is subject to substantial risks and uncertainties. Forward-Looking Statements
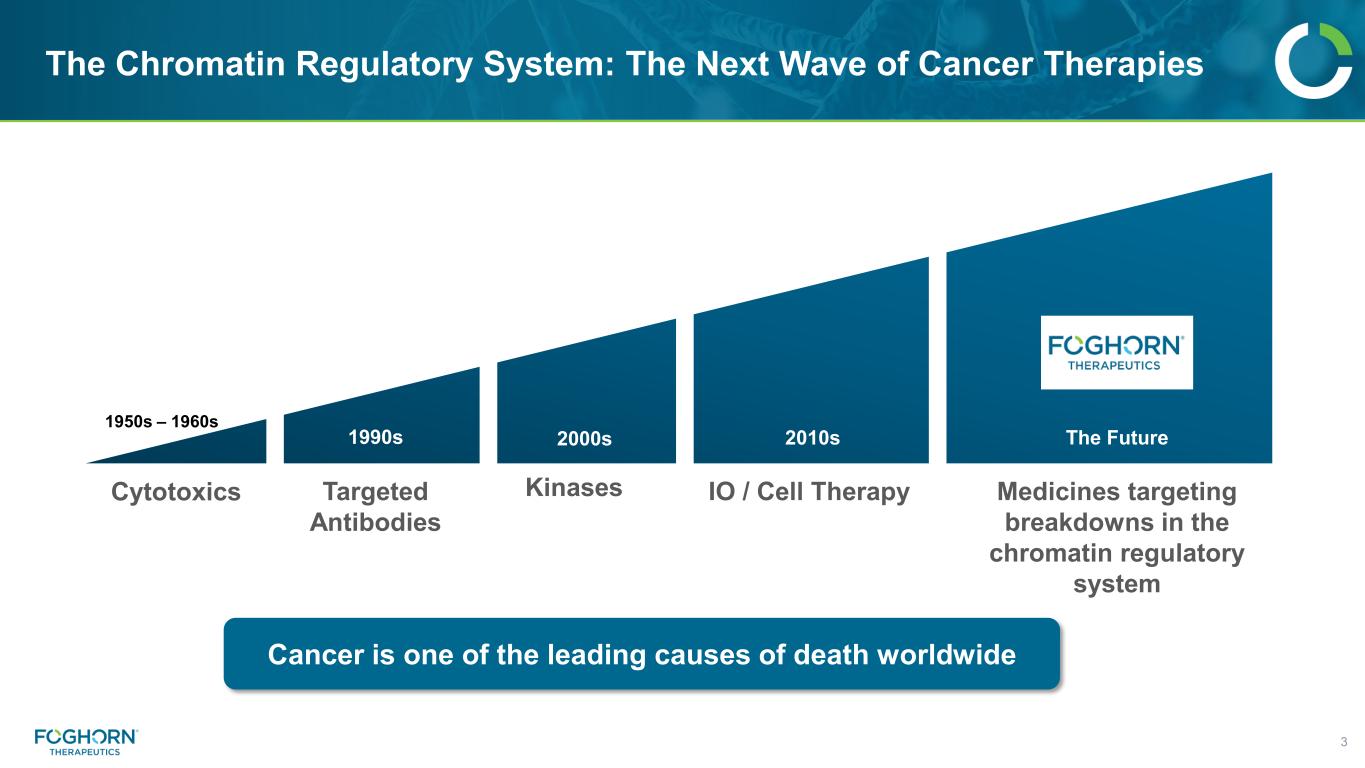
The Chromatin Regulatory System: The Next Wave of Cancer Therapies 3 Cytotoxics Targeted Antibodies IO / Cell Therapy Medicines targeting breakdowns in the chromatin regulatory system Cancer is one of the leading causes of death worldwide 1950s – 1960s Kinases 1990s 2000s 2010s The Future

Dysregulation of the Chromatin Regulatory System Has Been Implicated in up to 50% of All Cancers 4 Significant Market Opportunity 2.5M People 50% of All Cancers Based on exome sequencing, the chromatin regulatory system is implicated in ~50% of all cancers $400+ billion 2030 global oncology market opportunity Patients impacted by these cancers
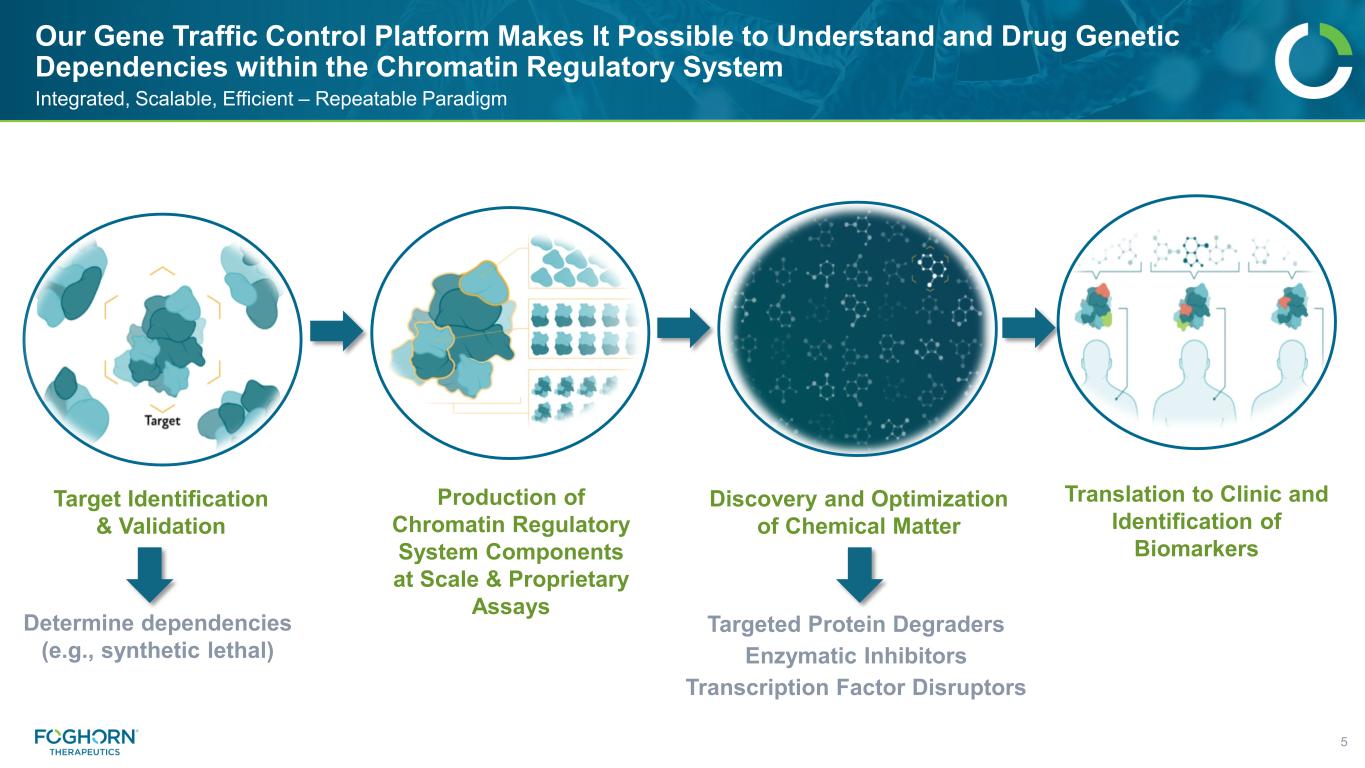
Our Gene Traffic Control Platform Makes It Possible to Understand and Drug Genetic Dependencies within the Chromatin Regulatory System 5 Integrated, Scalable, Efficient – Repeatable Paradigm Target Identification & Validation Production of Chromatin Regulatory System Components at Scale & Proprietary Assays Discovery and Optimization of Chemical Matter Targeted Protein Degraders Enzymatic Inhibitors Transcription Factor Disruptors Translation to Clinic and Identification of Biomarkers Determine dependencies (e.g., synthetic lethal)

Production of Chromatin Regulatory System Components Platform is Powered by Ability to Produce Components at Scale Drives Drug Discovery Pipeline with Cutting Edge Technology Surface Mapping Assembly HTS Biophysics/ SPR Affinity Screening and Validation Characterize TF/BAF Binding Sites Synthesize subcomplexes to enable drug discovery ASMS on full complex to yield novel degraders Multiple screening options with full complex Validation of novel small molecule binders BenefitsFeatures
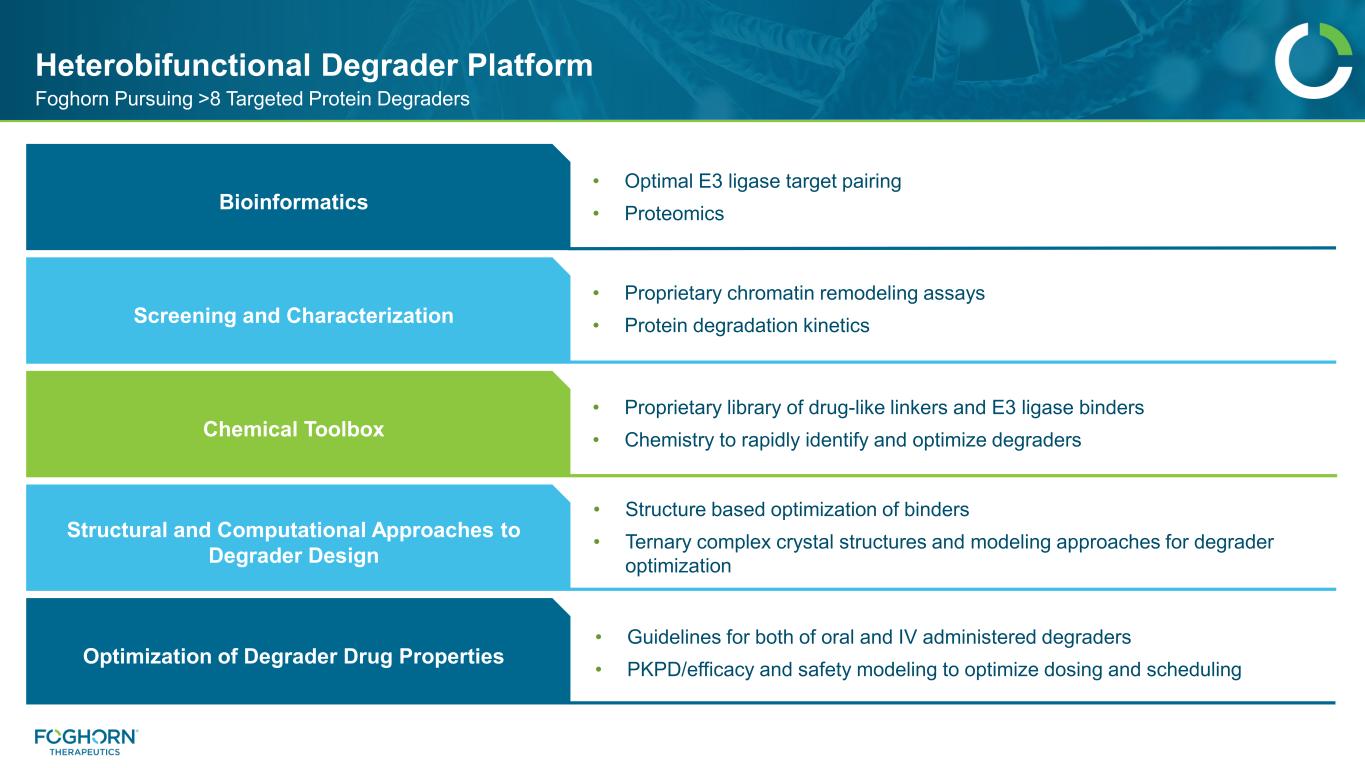
Foghorn Pursuing >8 Targeted Protein Degraders Heterobifunctional Degrader Platform Bioinformatics • Optimal E3 ligase target pairing • Proteomics Chemical Toolbox Screening and Characterization Structural and Computational Approaches to Degrader Design Optimization of Degrader Drug Properties • Proprietary chromatin remodeling assays • Protein degradation kinetics • Proprietary library of drug-like linkers and E3 ligase binders • Chemistry to rapidly identify and optimize degraders • Structure based optimization of binders • Ternary complex crystal structures and modeling approaches for degrader optimization • Guidelines for both of oral and IV administered degraders • PKPD/efficacy and safety modeling to optimize dosing and scheduling

Program / Target Modality Discovery IND-enabling Phase 1 Phase 2 Phase 3 Global Rights FHD-286 (BRG1 / BRM) Enzyme inhibitor FHD-609 (BRD9) Protein degrader Selective BRM I) Enzyme inhibitor II) Protein degrader Selective ARID1B Protein degrader Synthetic Lethal Targets (multiple) I) Enzyme inhibitors II) Protein degraders Transcription Factors (multiple) Transcription factor disruptors Partnered program (undisclosed) Transcription factor disruptor 8 Precision Oncology / Breadth and Depth First Two Programs in the Clinic, Broad Pipeline Advancing AML Early Clinical Data (Q4 2021) Uveal melanoma Early Clinical Data (Q4 2021) Synovial sarcoma Early Clinical Data (H1 2022) BRG1 mutated cancers IND 2022 ARID1A mutated cancers BRG1 mutated cancers Gene Traffic Control® Platform

The Chromatin Regulatory System Orchestrates Gene Expression
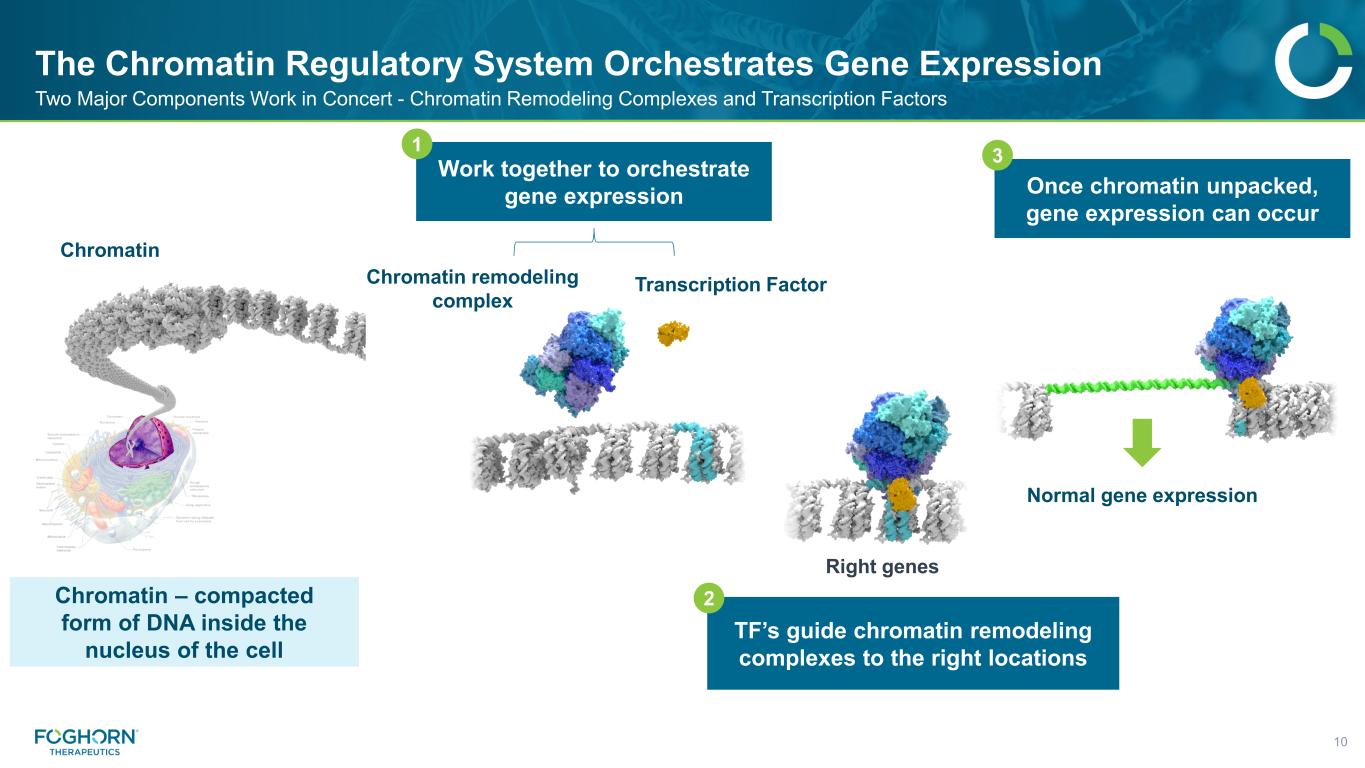
Normal gene expression The Chromatin Regulatory System Orchestrates Gene Expression 10 Two Major Components Work in Concert - Chromatin Remodeling Complexes and Transcription Factors Work together to orchestrate gene expression TF’s guide chromatin remodeling complexes to the right locations Once chromatin unpacked, gene expression can occur 1 2 3 Chromatin – compacted form of DNA inside the nucleus of the cell Chromatin Right genes Transcription FactorChromatin remodeling complex

Breakdowns in the Chromatin Regulatory System Lead to Disease 11 Mutations or overexpression in chromatin remodeling complexes result in abnormal gene expression Mutated or overexpressed TF hijacks chromatin remodeling complex to wrong location DISEASE DISEASE

Chromatin Regulatory System – Abundance of Targets 12 28 Chromatin Remodeling Complexes and >1,000 TFs Estimate >100 Transcription Factors Associated with just the BAF Complex BAF Complex and Associated Transcription Factors + BAF Complex Subunits Mutated and Dysregulated in Cancer

Mutations Lead to Disease Specific Genetic Dependencies on the Chromatin Regulatory System 13 Transcription Factor Mutations / Overexpression Chromatin Remodeling Complexes Mutations / Overexpression Mutations that Impinge on the Chromatin Regulatory System Novel Targets / Dependencies Targeted Protein Degradation: Bi-functional protein degraders for targets with no enzymatic activity Enzymatic Inhibitors: Highly selective and allosteric small molecule inhibitors Transcription Factor Disruptors: Disrupt interactions between chromatin remodeling complexes and transcription factors Tailored Drugging Approaches ATP ADP Potential druggable sites

FHD-286: Clinical Entry Point - AML and Uveal Melanoma FHD-286 is a Potent, Selective, Allosteric, Small Molecule Inhibitor of the BRG1 and BRM subunits of the BAF complex

FHD-286 Targets Abnormal Dependencies on BAF in Cancer • BRM/BRG1 is the engine (ATPase) of the BAF chromatin remodeling complex • BRG1 & BRM are highly similar proteins BRG and BRM Subunits BRMBRG1 Target / Approach • BRG1/BRM ATPase • Small molecule, allosteric, oral enzymatic inhibitor Indications • Acute myelogenous leukemia (AML) • Uveal melanoma • Indication expansion work ongoing in multiple solid tumors Mutation / Aberration • AML: Elevated BRG1-BAF / TF activity in AML blast cells • Uveal Melanoma: GNAQ/GNA11 mutated UM is driven by dependency on BAF / TF activity Program Status / Milestones • Phase I studies enrolling in AML and metastatic uveal melanoma • Phase I data as early as Q4’21 New Patients Impacted / Year* • AML: Over 20,000 relapsed and/or refractory patients • Uveal melanoma: Over 5,000 patients BAF Chromatin Remodeling Complex * US, EU5, Japan

AML & Dependency on BRG1 / Lineage Dependent TF Interactions Disease State Treatment with FHD-286 Loss of blast phenotype / Apoptosis Differentiation AML blasts stuck in BAF / TF dependent proliferative phase HSC Cancerous blast cells rely on BRG1 containing BAF / TF activity BAF – TF Activity BAF – TF Activity

AML Dependent on BRG1 / Lineage TF Interaction 17 Subtype Sample ID TF #1 SPI1 (PU.1) TF #3 TF #4 SPI1 (PU.1) / BAF Dependency SPI1 ChIPseq TF Association with AML by FAB Classification: 70% BRG1 Inhibition Leads to Loss of SPI1 (PU.1) Occupancy on Chromatin

FHD-286 Shows Broad Efficacy Across AML Patient Derived Samples Notable Patient ID Deep Response Pathology Review Disease Status 1690AML1 Y AML Secondary 1695AML1 Y AML/MDS Secondary 1696AML1 Y AML Secondary 1701AML1 Y AML Secondary 1893AML1 Y AML R/R 1899AML1 Y AML R/R 1990pAML1 Y AML R/R 1991pAML1 Y AML de novo 2041AML1 Y N/A de novo 2043pAML1 Y AML R/R 2059AML1 Y AML R/R 1682AML1 ~ N/A N/A 1689AML1 ~ AML/MDS de novo 1684AML1 N CML R/R 1924AML1 N AML/MDS R/R • Response observed in a majority of primary AML samples, irrespective of prior treatment or disease stage • Additional data set from patient derived samples demonstrate mutation agnostic responses 1695AML1 – BM-secondary AML Aza + venetoclax 7+3 Pro-differentiation effect Y = Deep reduction in blast cells ~ = Partial reduction N = No response
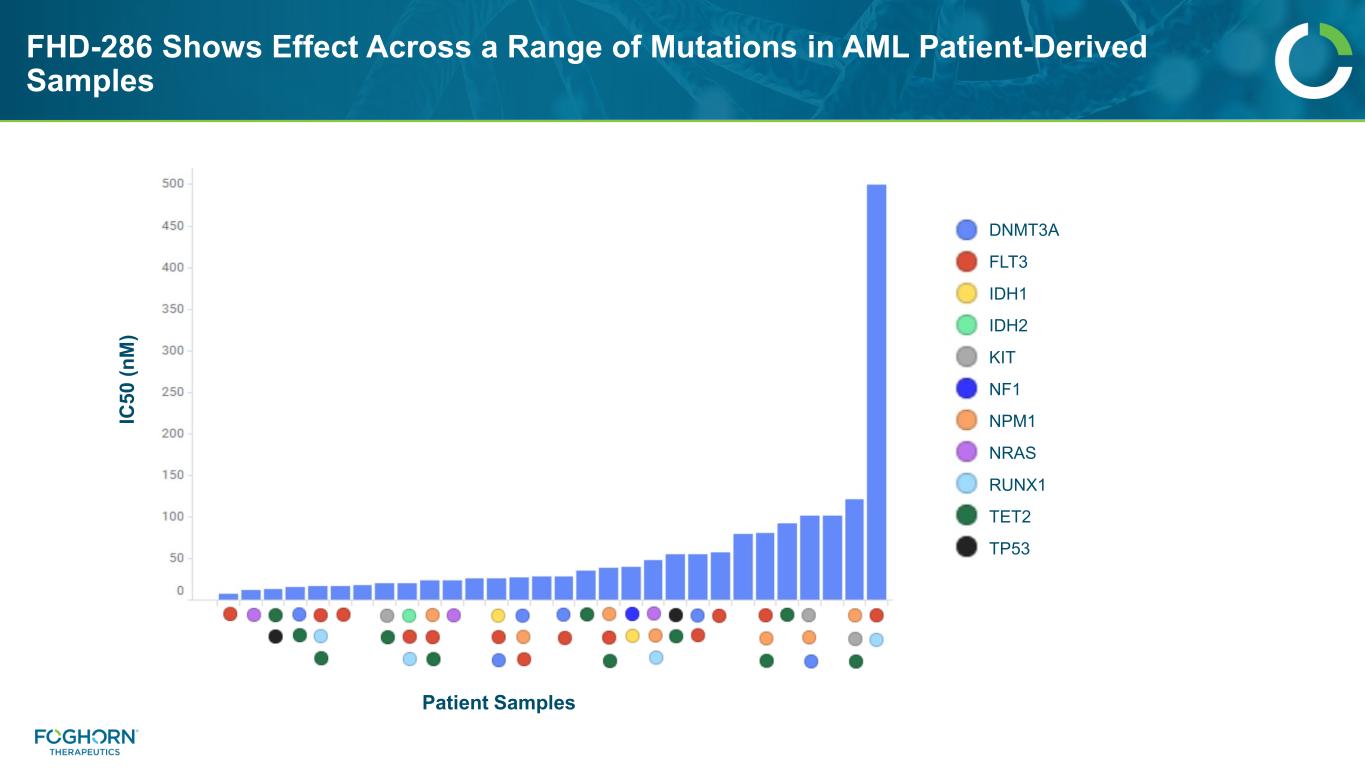
FHD-286 Shows Effect Across a Range of Mutations in AML Patient-Derived Samples IC 50 (n M ) Patient Samples DNMT3A FLT3 IDH1 IDH2 KIT NF1 NPM1 NRAS RUNX1 TET2 TP53

Dose-Dependent Tumor Growth Inhibition Observed with FHD-286 Treatment in AML CDX Models 20 Tumor Volume Body Weight 0 7 14 21 0 250 500 750 1000 1250 Days Post Treatment Tu m or V ol um e (m m 3 ) 0 7 14 21 0 800 1600 2400 3200 Days Post Treatment Tu m or V ol um e (m m 3 ) 0 7 14 21 80 90 100 110 120 Days Post Treatment Bo dy W ei gh t C ha ng e (% ) 0 7 14 21 80 90 100 110 120 Days Post Treatment B od y W ei gh t C ha ng e (% ) MV4-11 CDX Model (FLT3 ITD, MLL-AF4) OCI-AML2 CDX Model (MII-AF6, DNMT3a mut.) Tumor Volume Body Weight

Tumor Growth Inhibition with FHD-286 Treatment Observed by Bioluminescence Imaging in a Disseminated AML model FHD-286 1.5 mg/kg, BID (5on / 8off) x3 Sorafenib 15 mg/kg, QDx14 Vehicle FHD-286 1.5 mg/kg QDx28
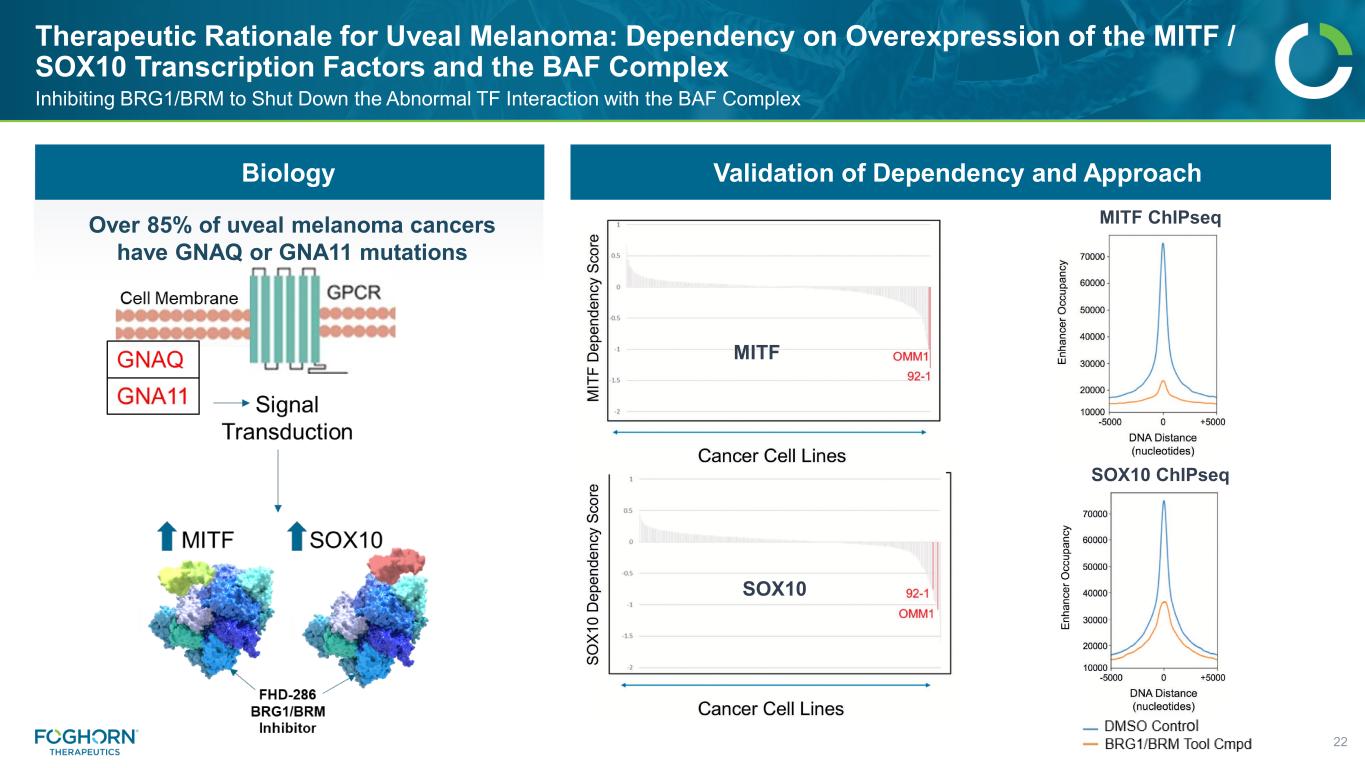
Therapeutic Rationale for Uveal Melanoma: Dependency on Overexpression of the MITF / SOX10 Transcription Factors and the BAF Complex 22 Inhibiting BRG1/BRM to Shut Down the Abnormal TF Interaction with the BAF Complex SOX10 MITF MITF ChIPseq SOX10 ChIPseq Biology Over 85% of uveal melanoma cancers have GNAQ or GNA11 mutations Validation of Dependency and Approach
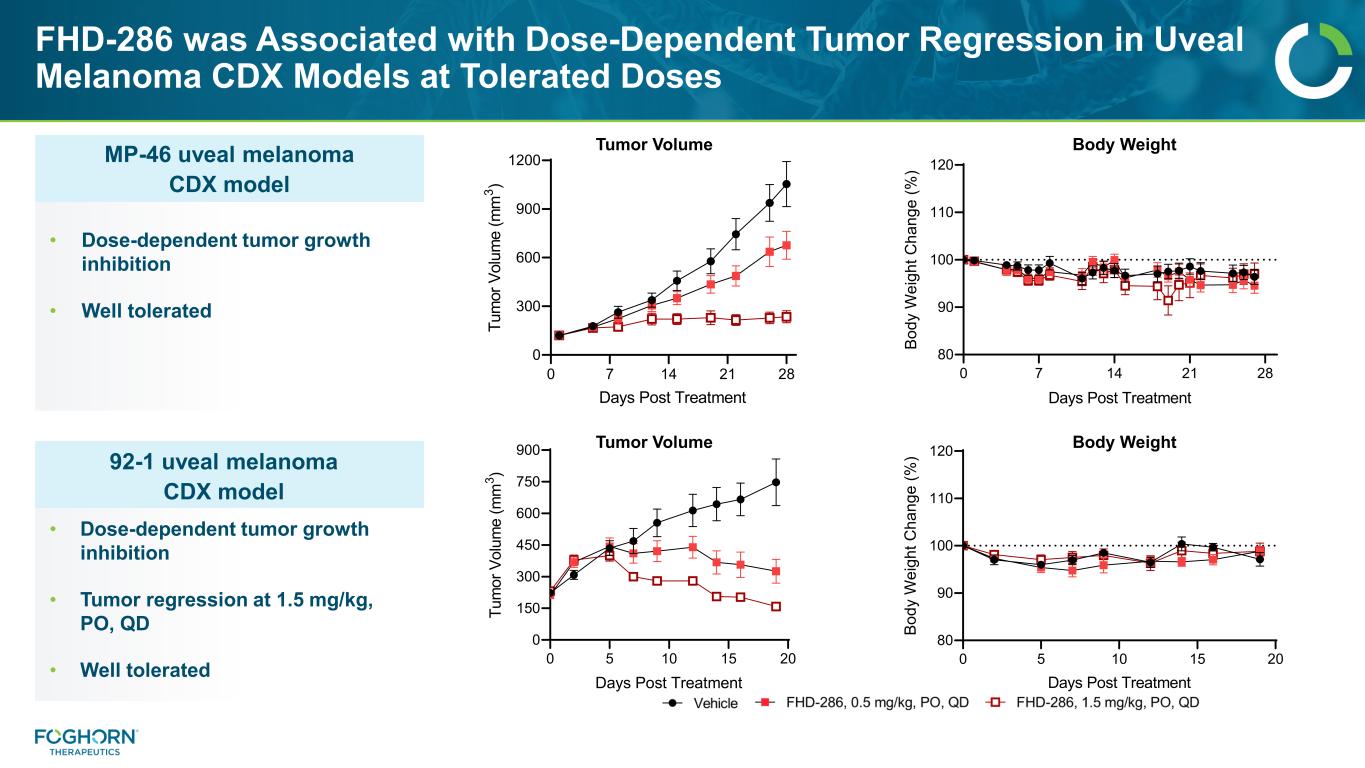
FHD-286 was Associated with Dose-Dependent Tumor Regression in Uveal Melanoma CDX Models at Tolerated Doses 0 7 14 21 28 0 300 600 900 1200 Days Post Treatment Tu m or V ol um e (m m 3 ) Tumor Volume 0 7 14 21 28 80 90 100 110 120 Days Post Treatment Bo dy W ei gh t C ha ng e (% ) Body Weight 0 5 10 15 20 0 150 300 450 600 750 900 Days Post Treatment Tu m or V ol um e (m m 3 ) 0 5 10 15 20 80 90 100 110 120 Days Post Treatment Bo dy W ei gh t C ha ng e (% ) Tumor Volume Body Weight MP-46 uveal melanoma CDX model • Dose-dependent tumor growth inhibition • Well tolerated 92-1 uveal melanoma CDX model • Dose-dependent tumor growth inhibition • Tumor regression at 1.5 mg/kg, PO, QD • Well tolerated

FHD-286 Clinical Development Plan Two Parallel Phase 1 Studies Activated Clinical data as early as Q4 2021 Trial Designs • Single patient accelerated titration (n=1) • Convert to 3+3 once relevant PK/PD, safety or clinical activity observed • Retrospective biomarker analysis to further evaluate safety and efficacy • Assess safety, PK, biomarkers and efficacy CLINICAL PLAN AML & Uveal Melanoma FIH Phase 1 Studies Expansion cohorts in AML, UM and potentially other indications Potential for entry into definitive efficacy trials in AML Potential for entry into definitive efficacy trials in metastatic uveal melanoma Potential for Indication Expansion Beyond AML and UM Metastatic Uveal Melanoma Relapsed / Refractory AML & MDS

FHD-609: Clinical Entry Point – Synovial Sarcoma FHD-609 is a Selective, Potent, Protein Degrader of the BRD9 component of the BAF complex

BRD9 is required for the survival of synovial sarcoma cells FHD-609 Targets and Degrades the BRD9 subunit of BAF which is Required for Synovial Sarcoma Cells to Survive Selective, Potent BRD9 Targeted Protein Degrader SS18-SSX1 / SSX2 / SSX4 mutated subunit Target / Approach • BRD9 • Intravenous Protein Degrader Initial Indication • Synovial Sarcoma Mutation / Aberration • SS18-SSX1 / SSX2 / SSX4 protein fusions Program Status / Milestones • Phase I data as early as H1’22 New Patients Impacted / Year* • Synovial Sarcoma: Over 1,800 patients / year B R D 9 D ep en de nc y Sc or e BRD9 subunit * US, EU5, Japan

Robust in vivo Activity Observed in Synovial Sarcoma Model and BRD9 Degradation Associated with FHD-609 Treatment Weekly Dosing of FHD-609 Achieved Sustained BRD9 Degradation SY01 Synovial Sarcoma CDX Model • Mutation: SS18-SSX2 • Inhibited tumor growth • Dose dependent BRD9 degradation correlated with anti-tumor activity Sustained BRD9 Degradation
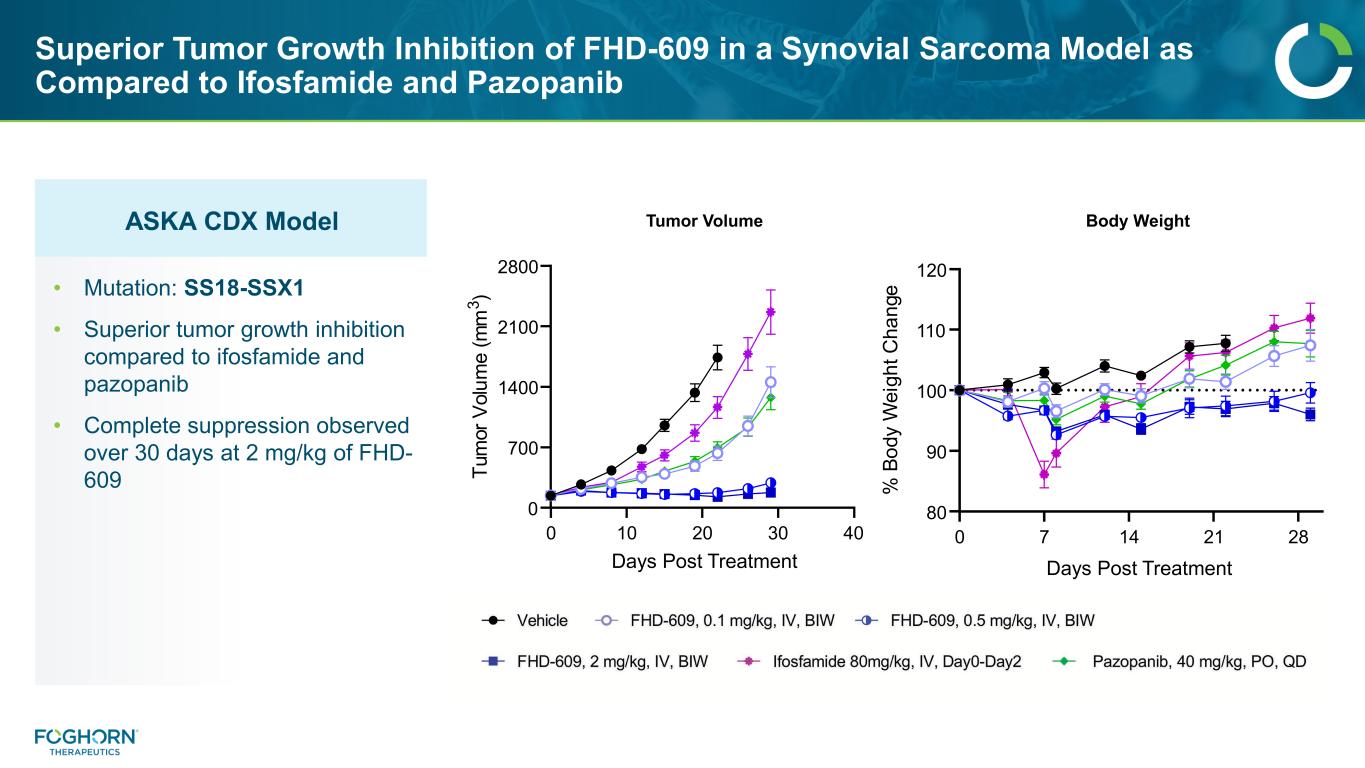
ASKA CDX Model Superior Tumor Growth Inhibition of FHD-609 in a Synovial Sarcoma Model as Compared to Ifosfamide and Pazopanib Tumor Volume Body Weight • Mutation: SS18-SSX1 • Superior tumor growth inhibition compared to ifosfamide and pazopanib • Complete suppression observed over 30 days at 2 mg/kg of FHD- 609 0 7 14 21 28 80 90 100 110 120 Days Post Treatment % Bo d y W ei gh t C ha ng e 0 10 20 30 40 0 700 1400 2100 2800 Days Post Treatment Tu m or Vo lu m e (m m 3 )

ASKA CDX Model Superior Tumor Growth Inhibition of FHD-609 in a Synovial Sarcoma Model as Compared to Ifosfamide and Pazopanib 29 0 10 20 30 40 0 700 1400 2100 2800 Days Post Treatment Tu m or V ol um e (m m 3 ) 0 7 14 21 28 80 90 100 110 120 Days Post Treatment % B od y W ei gh t C ha ng e Tumor Volume Body Weight • Mutation: SS18-SSX1 • Superior tumor growth inhibition compared to ifosfamide and pazopanib • Complete suppression observed over 30 days at 2 mg/kg of FHD- 609

FHD-609 Clinical Development Plan Metastatic Synovial Sarcoma Synovial Sarcoma expansion cohorts Synovial Sarcoma FIH Phase 1 Potential for entry into definitive efficacy trials in synovial sarcoma CLINICAL PLAN Clinical data as early as H1 2022 SMARCB-1 deleted tumors and potentially other indications Trial Designs • Single patient accelerated titration (n=1) • Convert to 3+3 once relevant PK/PD, safety or clinical activity observed • Assess safety, PK, clinical activity and biomarkers Biomarkers: • SS18-SSX1, SS18-SSX2 or SS18-SSX4 translocation

Selective BRM Modulators for BRG1 Mutated Cancers Enzymatic Inhibitor and Protein Degrader Programs

BRG1 Mutations Create a Genetic Dependency on BRM Selective BRM Modulators Overview BRM BRG1 Target / Approach • BRM • Enzymatic inhibitor • Targeted protein degrader Indication • BRG1 mutated cancers (e.g., NSCLC), 30+ cancers with BRG1 mutations Mutation / Aberration • BRG1 Stage • Pre-clinical New Patients Impacted / year* • > 100,000 B R M D ep en de nc y Sc or e * US, EU5, Japan

BRG1 Mutated in ~5% of All Tumors Broad Addressable Patient Population BRG1 mutated across range of tumors Accounts for ~5% of all tumors BRG1 mutated in up to 10% of NSCLC tumors, minimal overlap with other mutations
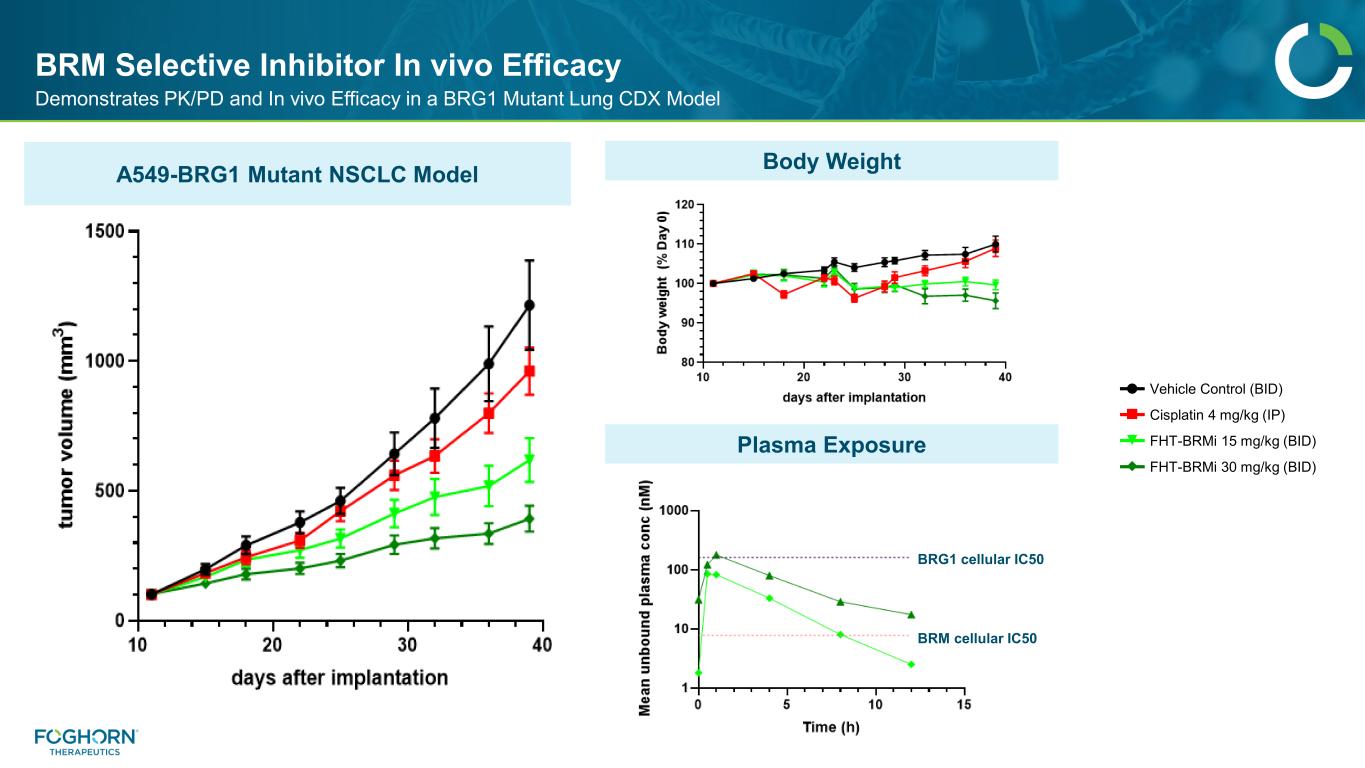
BRM Selective Inhibitor In vivo Efficacy Demonstrates PK/PD and In vivo Efficacy in a BRG1 Mutant Lung CDX Model BRG1 cellular IC50 BRM cellular IC50 A549-BRG1 Mutant NSCLC Model Body Weight Plasma Exposure Vehicle Control (BID) Cisplatin 4 mg/kg (IP) FHT-BRMi 15 mg/kg (BID) FHT-BRMi 30 mg/kg (BID)

Enzymatic selectivity approaching 200x achieved 35Company Confidential & Proprietary Information
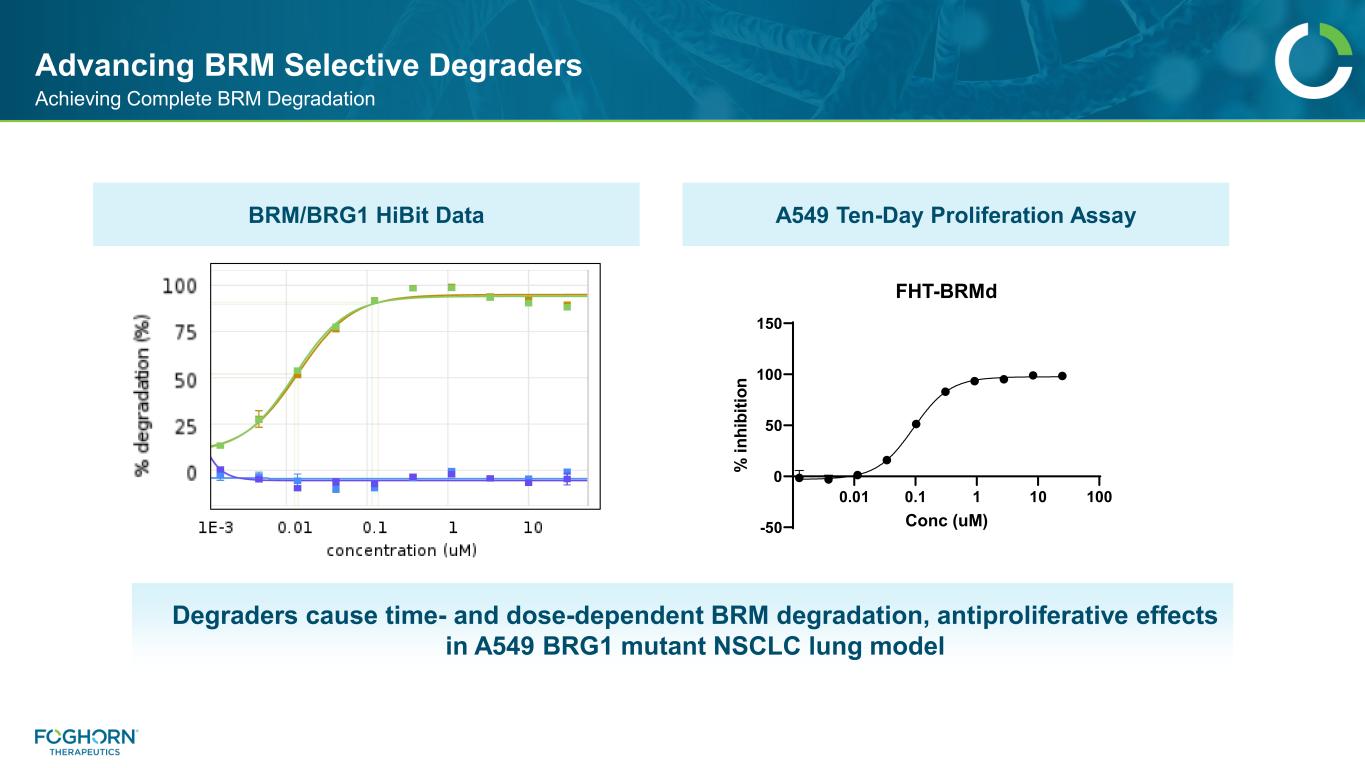
Advancing BRM Selective Degraders Achieving Complete BRM Degradation Degraders cause time- and dose-dependent BRM degradation, antiproliferative effects in A549 BRG1 mutant NSCLC lung model 0.01 0.1 1 10 100 -50 0 50 100 150 FHX-NSXLX Conc (uM) % in hi bi tio n F T-BRMd BRM/BRG1 HiBit Data A549 Ten-Day Proliferation Assay

Selective ARID1B Protein Degrader for ARID1A Mutated Cancers

ARID1A – Most Mutated Subunit in BAF Complex – Creates Dependency on ARID1B 38 Selective ARID1B Protein Degrader Overview ARID1A ARID1B Target / Approach • ARID1B • Targeted Protein Degrader Indication • ARID1A mutated cancers Mutation / Aberration • ARID1A mutations (e.g., ovarian, endometrial, colorectal, bladder and other cancers) Stage • Pre-clinical New Patients Impacted / year* • > 175,000 AR ID 1B D ep en de nc y Sc or e * US, EU5, Japan
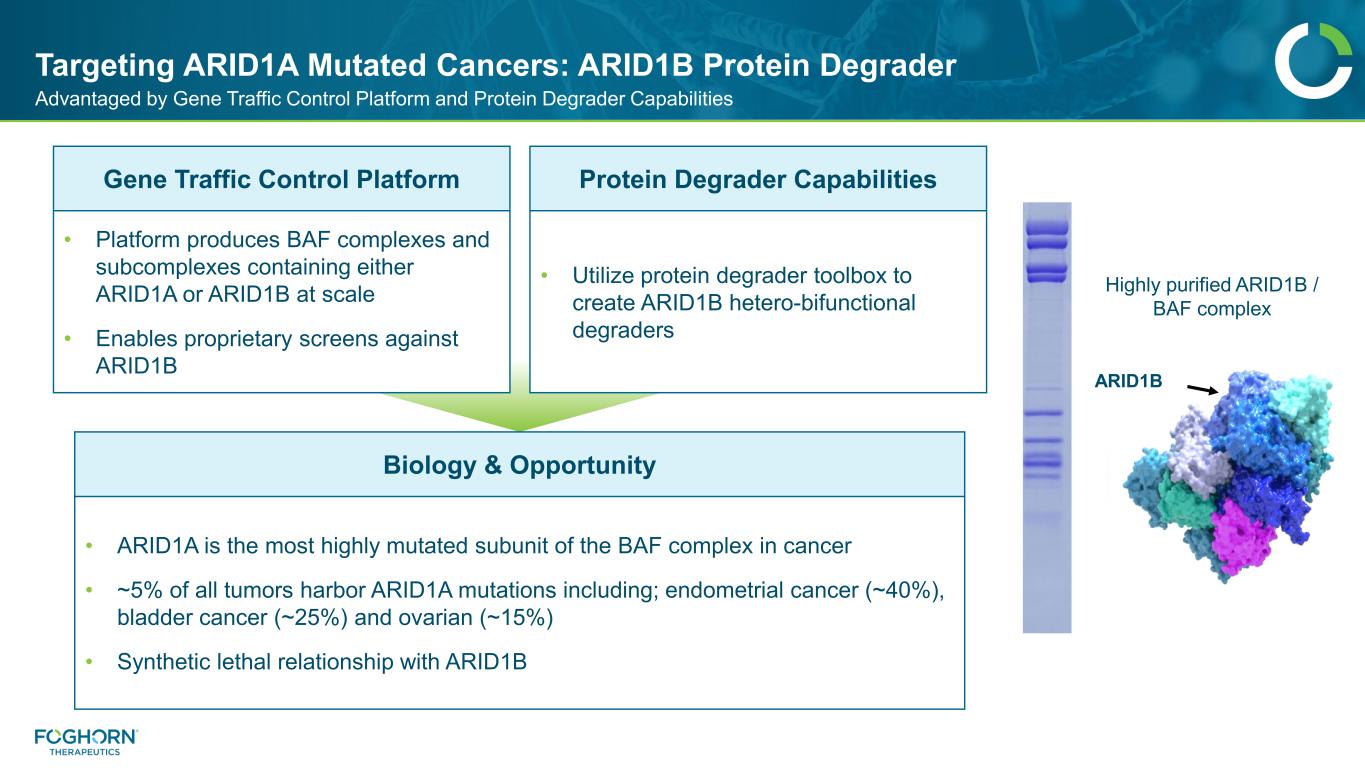
Targeting ARID1A Mutated Cancers: ARID1B Protein Degrader Advantaged by Gene Traffic Control Platform and Protein Degrader Capabilities Gene Traffic Control Platform Protein Degrader Capabilities • Platform produces BAF complexes and subcomplexes containing either ARID1A or ARID1B at scale • Enables proprietary screens against ARID1B • Utilize protein degrader toolbox to create ARID1B hetero-bifunctional degraders Biology & Opportunity • ARID1A is the most highly mutated subunit of the BAF complex in cancer • ~5% of all tumors harbor ARID1A mutations including; endometrial cancer (~40%), bladder cancer (~25%) and ovarian (~15%) • Synthetic lethal relationship with ARID1B ARID1B Highly purified ARID1B / BAF complex

Novel Approach to Targeting Transcription Factors Disrupting Transcription Factor – Chromatin Remodeling Complex Interactions

A New Approach to Drugging Transcription Factors Enabled by Proprietary Ability to Purify and Synthesize Chromatin Regulatory System Components 41 • Highly involved in gene expression • Implicated in range of cancers and other diseases TFs are compelling drug targets… • Featureless surface: no druggable binding pocket • Tight interactions with DNA: undruggable affinities …but historically difficult to target Historical Focus Potential druggable sites Foghorn’s Focus Foghorn has a new approach focusing on interaction with BAF Druggable binding pockets Druggable affinities

Transcription Factor-Chromatin Remodeling Complex Interactions 42 Unique Insights in Where and How Transcription Factors Bind TF #2TF #1 TF #3 Transcription Factors (TF): TF #4 0.01 0.1 1 10 100 1000 0 100 200 300 400 SPI1 (nM) RU KD = 21 nM 0.1 1 10 100 1000 10000 0 50 100 150 200 250 MYOD1 (nM) RU KD = 125 nM KD = 351 nM 0.1 1 10 100 1000 10000 0 50 100 150 200 250 300 350 MITF (nM) RU KD = 94 nM TF #1 (nM) TF #2 (nM) TF #3 (nM) TF #4 (nM)

• Merck collaboration to drug single specified transcription factor target • $425 million in up-front, research, development and sales-based milestones • Up to low double-digit royalties on product sales Highly Scalable Approach and Significant Unmet Medical Need 43 Potential to Drug > 100 TFs Associated with BAF • >100 TFs estimated associated with BAF • Foghorn pursuing multiple TFs in parallel • Approach highly scalable and potential broad application – other chromatin remodeling complexes and other diseases
Investment Highlights LARGE MARKET POTENTIAL Biology implicated in up to 50% of cancer potentially impacting ~2.5 million patients Potential applications beyond oncology in diseases including virology, autoimmune disease and neurology 44 WELL FUNDED MEANINGFUL UPCOMING MILESTONES Phase I FHD-286 data as early as Q4’21 Phase I FHD-609 data as early as H1’22 $160.9 million cash and equivalents as of 3/31/2021 EXPERIENCED LEADERSHIP TEAM Expertise across drug discovery, clinical development and commercialization Over 220 drug candidates into the clinic and over 30 drugs approved

Appendix

Carl Decicco, Ph.D., CSO Proven Leadership Team 46 Steve Bellon, Ph.D., SVP, Drug DiscoveryAdrian Gottschalk, President & CEO Carlos Costa, SVP, HR Sam Agresta, M.D., M.P.H., CMO Jacqueline Cinicola, VP Regulatory Affairs Murphy Hentemann, Ph.D., VP Program Leadership Chong-Hui Gu, VP, CMC and QA Ryan Kruger, PhD, VP, Biology Fanny Cavalie, SVP, Business & Operations David Millan, Ph.D, VP, Chemistry Michael LaCascia, CLO Nicola Majchrzak, VP, Clinical DevelopmentAllan Reine, M.D., CFO Scott Innis, VP, Program Leadership

Charles Sawyers, M.D. MSKCC, HHMI – SAB Chair Gerald Crabtree, M.D. Stanford, HHMI; Founder Faheem Hasnain Gossamer Bio, Chair of Mirati David Schenkein, M.D. General Partner, GV Craig Peterson, Ph.D. Professor UMass Medical School Tony Kouzarides, Ph.D. Gurdon Institute – University of Cambridge Doug Cole, M.D. Flagship Pioneering – Board Chair; Founder Cigall Kadoch, Ph.D. Dana-Farber, Broad, HMS; Founder Scott Biller, Ph.D. Former CSO and Strategic Advisor, Agios Adam Koppel, M.D., Ph.D. Bain Capital Life Sciences Simba Gill, Ph.D. Evelo Biosciences, Partner at Flagship Pioneering Michael Mendelsohn, M.D. Cardurion Pharmaceuticals Adrian Gottschalk Foghorn President & CEO Ian Smith Exec. Chair of Solid Bio., Chair of ViaCyte, Former COO of Vertex Experienced Leadership Team with Industry Leading Advisors and Investors 47 BOARD OF DIRECTORS SCIENTIFIC AND OTHER ADVISORS
