Attached files
| file | filename |
|---|---|
| EX-99.2 - EX-99.2 - Wave Life Sciences Ltd. | d324054dex992.htm |
| 8-K - 8-K - Wave Life Sciences Ltd. | d324054d8k.htm |

Corporate Presentation January 6, 2017 Exhibit 99.1

Forward Looking Statements This document contains forward-looking statements. All statements other than statements of historical facts contained in this document, including statements regarding possible or assumed future results of operations, business strategies, development plans, regulatory activities, competitive position, potential growth opportunities, use of proceeds and the effects of competition are forward-looking statements. These statements involve known and unknown risks, uncertainties and other important factors that may cause the actual results, performance or achievements of WAVE Life Sciences Ltd. (the “Company”) to be materially different from any future results, performance or achievements expressed or implied by the forward-looking statements. In some cases, you can identify forward-looking statements by terms such as “may,” “will,” “should,” “expect,” “plan,” “aim,” “anticipate,” “could,” “intend,” “target,” “project,” “contemplate,” “believe,” “estimate,” “predict,” “potential” or “continue” or the negative of these terms or other similar expressions. The forward-looking statements in this presentation are only predictions. The Company has based these forward-looking statements largely on its current expectations and projections about future events and financial trends that it believes may affect the Company’s business, financial condition and results of operations. These forward-looking statements speak only as of the date of this presentation and are subject to a number of risks, uncertainties and assumptions, including those listed under Risk Factors in the Company’s Form 10-K and other filings with the SEC, some of which cannot be predicted or quantified and some of which are beyond the Company’s control. The events and circumstances reflected in the Company’s forward-looking statements may not be achieved or occur, and actual results could differ materially from those projected in the forward-looking statements. Moreover, the Company operates in a dynamic industry and economy. New risk factors and uncertainties may emerge from time to time, and it is not possible for management to predict all risk factors and uncertainties that the Company may face. Except as required by applicable law, the Company does not plan to publicly update or revise any forward-looking statements contained herein, whether as a result of any new information, future events, changed circumstances or otherwise.

WAVE Life Sciences Overview Genetic medicines company developing targeted nucleic acid therapies for patients impacted by rare diseases Rationally designed nucleic acid therapeutics optimized by stereochemistry Three lead candidates initiating clinical trials in 2017 Six development programs by end of 2018 Core focus and expertise in neurological rare genetic diseases Strategic partnerships for non-core assets, addressing multiple therapeutic areas Proprietary R&D platform and advanced chemistry: a sustainable engine for future growth Cash runway into 2019
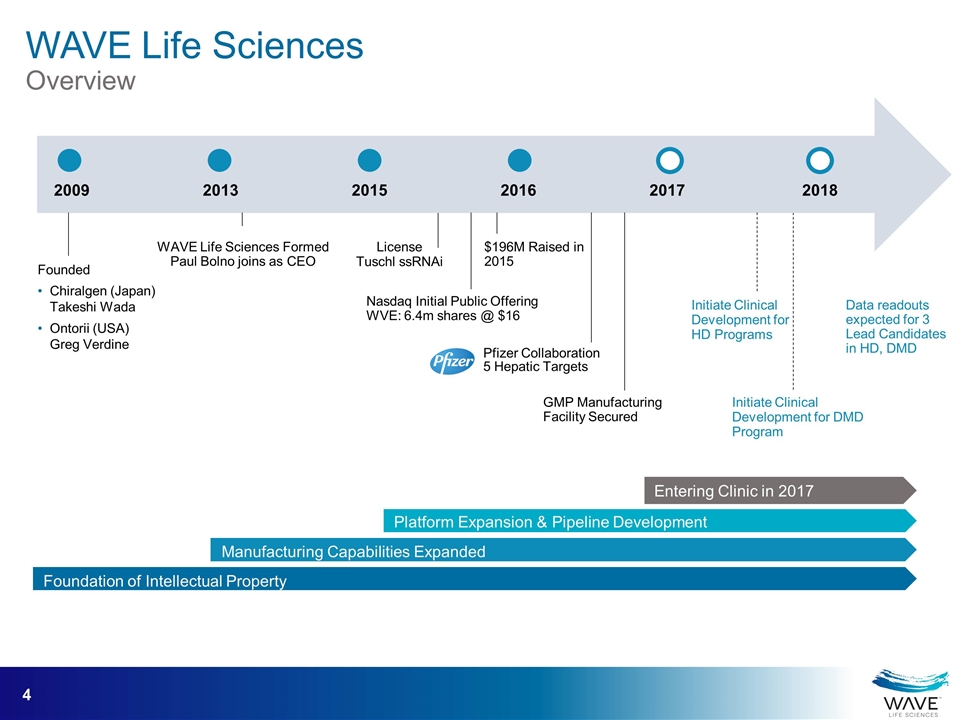
Founded Chiralgen (Japan) Takeshi Wada Ontorii (USA) Greg Verdine 2009 2013 WAVE Life Sciences Formed Paul Bolno joins as CEO 2015 Nasdaq Initial Public Offering WVE: 6.4m shares @ $16 License Tuschl ssRNAi 2016 2017 2018 Initiate Clinical Development for HD Programs Data readouts expected for 3 Lead Candidates in HD, DMD Foundation of Intellectual Property Manufacturing Capabilities Expanded Platform Expansion & Pipeline Development Entering Clinic in 2017 Pfizer Collaboration 5 Hepatic Targets GMP Manufacturing Facility Secured Initiate Clinical Development for DMD Program $196M Raised in 2015 WAVE Life Sciences Overview
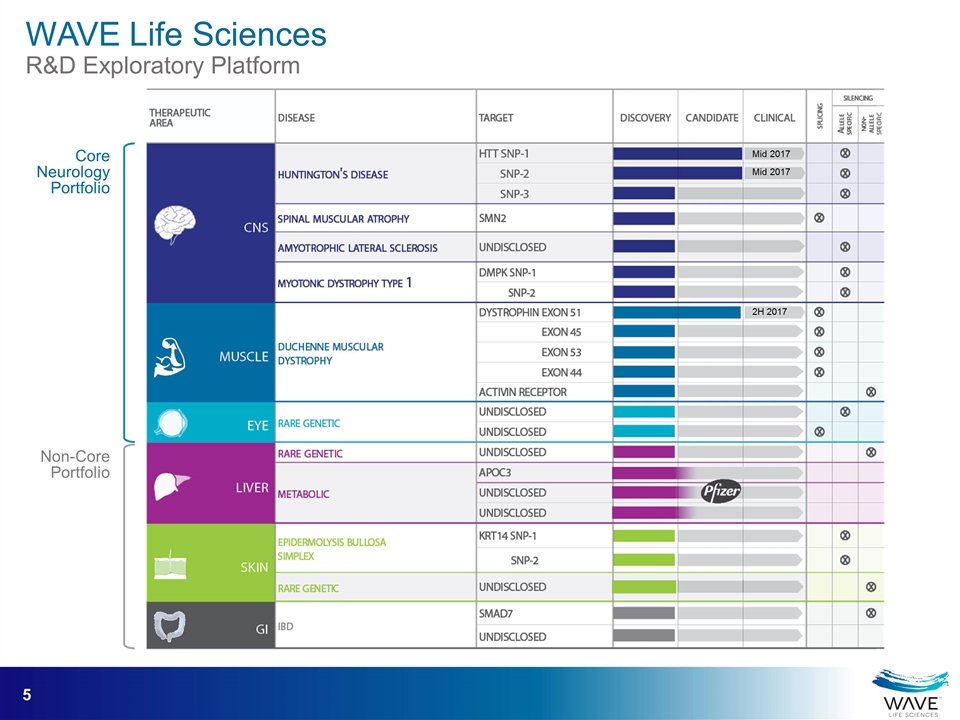
WAVE Life Sciences R&D Exploratory Platform Core Neurology Portfolio Mid 2017 Mid 2017 2H 2017 Non-Core Portfolio
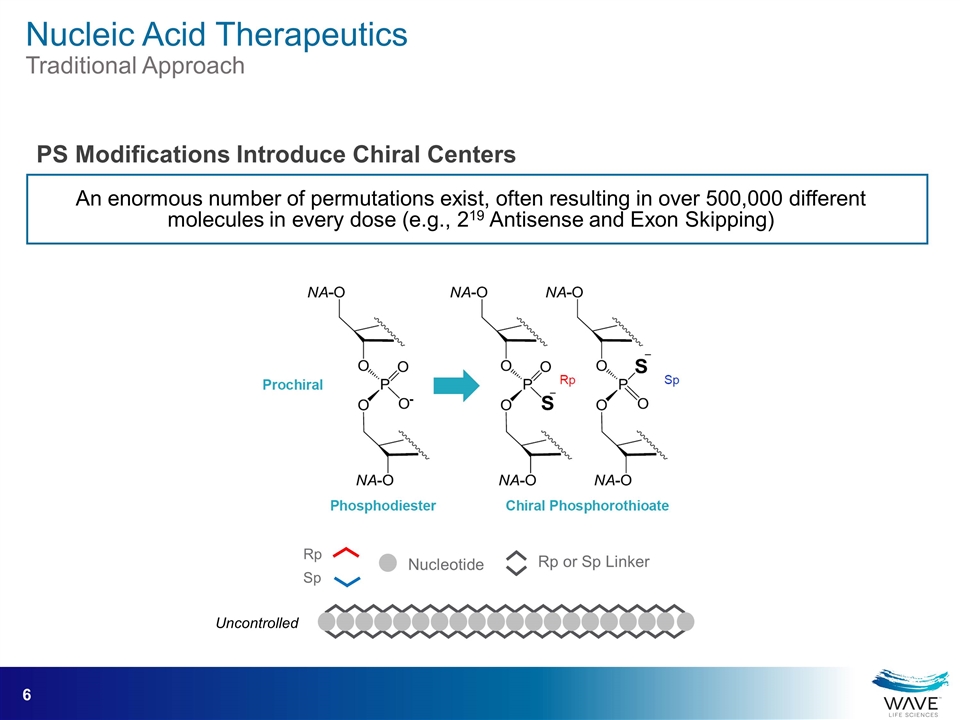
An enormous number of permutations exist, often resulting in over 500,000 different molecules in every dose (e.g., 219 Antisense and Exon Skipping) Rp Sp Rp or Sp Linker Nucleotide Uncontrolled PS Modifications Introduce Chiral Centers Nucleic Acid Therapeutics Traditional Approach
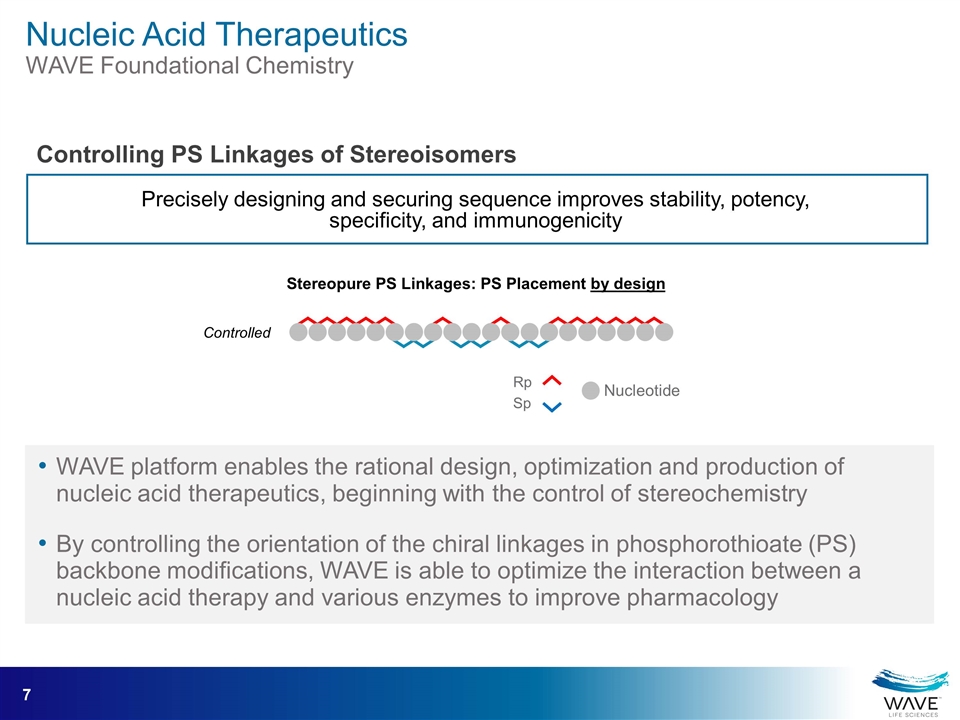
WAVE platform enables the rational design, optimization and production of nucleic acid therapeutics, beginning with the control of stereochemistry By controlling the orientation of the chiral linkages in phosphorothioate (PS) backbone modifications, WAVE is able to optimize the interaction between a nucleic acid therapy and various enzymes to improve pharmacology Precisely designing and securing sequence improves stability, potency, specificity, and immunogenicity Stereopure PS Linkages: PS Placement by design Controlled Rp Sp Nucleotide Nucleic Acid Therapeutics WAVE Foundational Chemistry Controlling PS Linkages of Stereoisomers
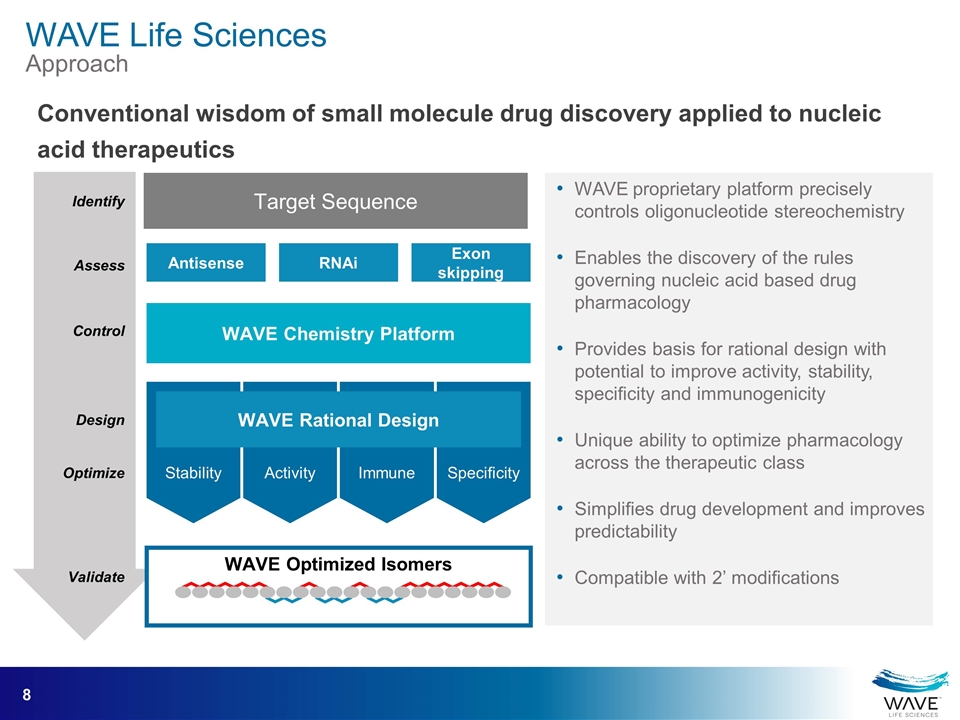
WAVE Optimized Isomers Stability Activity Immune Specificity WAVE Rational Design Antisense RNAi Exon skipping Target Sequence WAVE proprietary platform precisely controls oligonucleotide stereochemistry Enables the discovery of the rules governing nucleic acid based drug pharmacology Provides basis for rational design with potential to improve activity, stability, specificity and immunogenicity Unique ability to optimize pharmacology across the therapeutic class Simplifies drug development and improves predictability Compatible with 2’ modifications Conventional wisdom of small molecule drug discovery applied to nucleic acid therapeutics Assess Control Optimize Validate Design Identify WAVE Chemistry Platform WAVE Life Sciences Approach
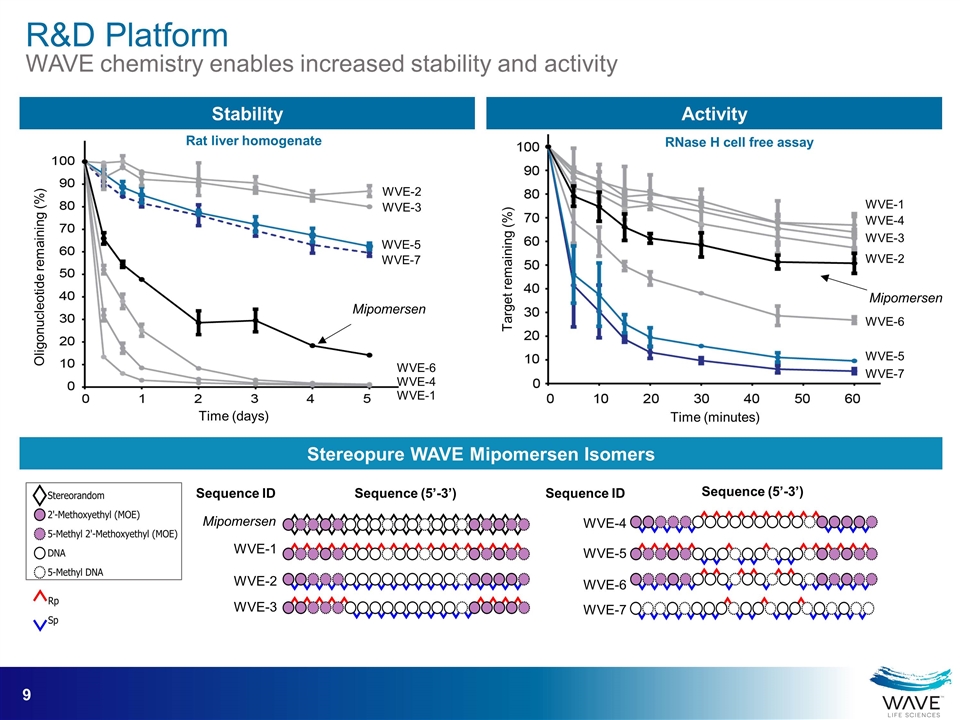
Mipomersen WVE-2 WVE-3 WVE-5 WVE-7 WVE-6 WVE-4 WVE-1 Oligonucleotide remaining (%) Time (days) Rat liver homogenate Stability Activity WVE-5 WVE-7 WVE-3 Mipomersen WVE-4 WVE-6 WVE-2 WVE-1 Sequence ID Sequence (5’-3’) Mipomersen WVE-6 WVE-4 WVE-1 WVE-2 WVE-3 WVE-5 WVE-7 Target remaining (%) Time (minutes) RNase H cell free assay Sequence ID Sequence (5’-3’) Stereopure WAVE Mipomersen Isomers R&D Platform WAVE chemistry enables increased stability and activity
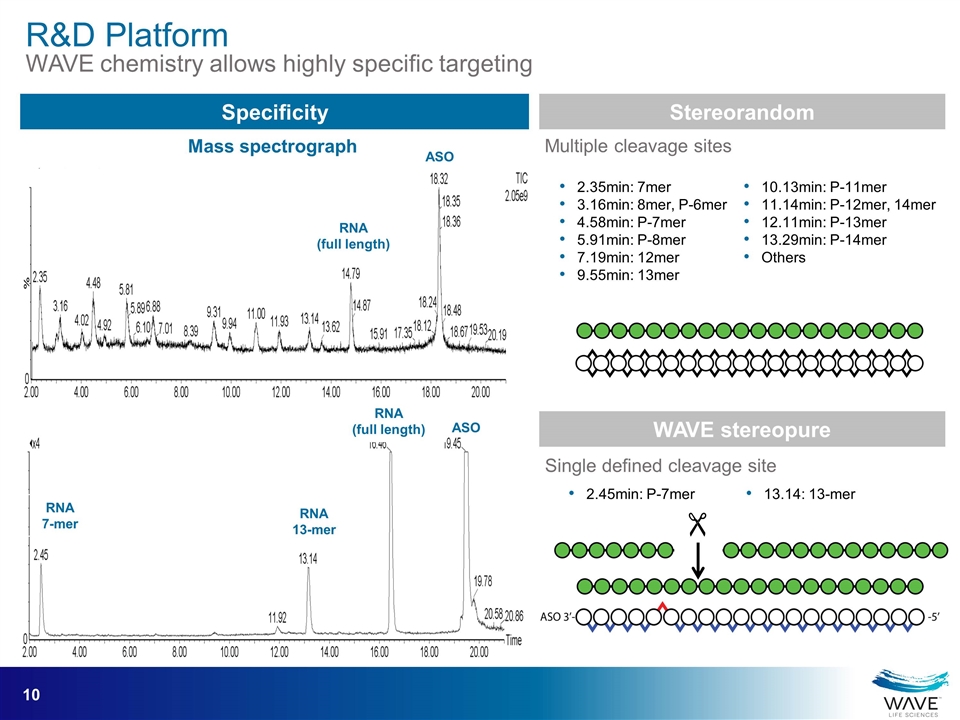
Specificity RNA (full length) ASO RNA 13-mer RNA 7-mer RNA (full length) ASO 2.35min: 7mer 3.16min: 8mer, P-6mer 4.58min: P-7mer 5.91min: P-8mer 7.19min: 12mer 9.55min: 13mer 10.13min: P-11mer 11.14min: P-12mer, 14mer 12.11min: P-13mer 13.29min: P-14mer Others 2.45min: P-7mer 13.14: 13-mer Stereorandom WAVE stereopure Multiple cleavage sites Single defined cleavage site " Mass spectrograph R&D Platform WAVE chemistry allows highly specific targeting
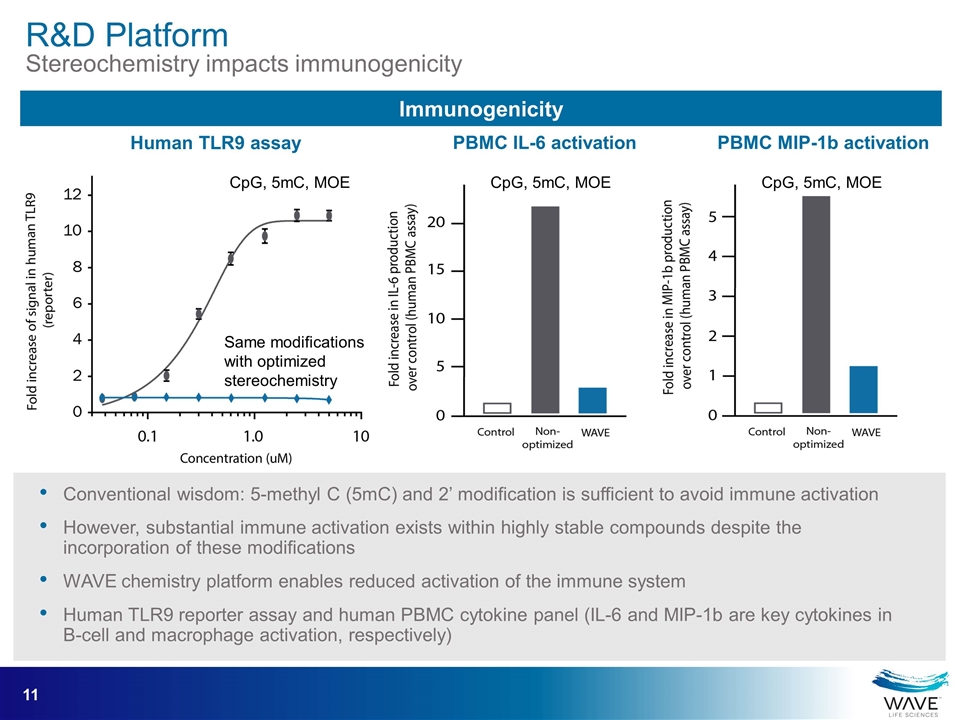
Immunogenicity CpG, 5mC, MOE Same modifications with optimized stereochemistry CpG, 5mC, MOE CpG, 5mC, MOE Human TLR9 assay PBMC IL-6 activation PBMC MIP-1b activation Conventional wisdom: 5-methyl C (5mC) and 2’ modification is sufficient to avoid immune activation However, substantial immune activation exists within highly stable compounds despite the incorporation of these modifications WAVE chemistry platform enables reduced activation of the immune system Human TLR9 reporter assay and human PBMC cytokine panel (IL-6 and MIP-1b are key cytokines in B-cell and macrophage activation, respectively) R&D Platform Stereochemistry impacts immunogenicity

WAVE Stereochemistry Patents Granted and Pending Foundational Chemistry Chiral Control and Design Platform Candidate Specific IP Co-exclusive license holder for single-strand RNAi (ssRNAi) patent estate Martinez J, Patkaniowska A, Urlaub H, Lührmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. License includes 19-29 nucleotides molecules; RNAi mechanism; various chemical modifications License to Pfizer hepatic targeting technology WAVE Intellectual Property
In June 2015, WAVE became the co-exclusive license holder (with Ionis Pharmaceuticals) to the single-strand RNAi patent estate (Tuschl, Zamore) Includes 19-29 nucleotides molecules; RNAi mechanism; various chemical modifications Single-stranded RNAi requires different design rules from double-stranded RNAi formats When properly optimized, single-stranded RNAi has comparable or greater potency than double-stranded RNAi formats Stereopure modifications are expected to provide further enhancements to pharmacology Martinez J, Patkaniowska A, Urlaub H, Lührmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. “Considering the feasibility of modulating the stability and uptake properties of single-stranded RNAs, 5′-phosphorylated single-stranded antisense siRNAs may further expand the utility of RNAi-based gene silencing technology as tool for functional genomics as well as therapeutic applications.” Single Strand RNAi New modalities
Continue to meet demand IND enabling studies GMP clinical trial supply Comparable cost of goods Potential cost-per-patient advantages (dose and schedule) Scalable Synthesis Superior Pharmacology Applicable Across Modalities Rational design of single drugs Increased activity Increased stability Increased specificity Reduced immune activation Compatible with targeting moieties (GalNAc, others) Other areas in development Antisense (RNase H) Exon skipping RNAi single-strand (Ago2) Guide-strand (CRISPR) Genetic medicines toolkit Other areas in development WAVE Advantages Summary
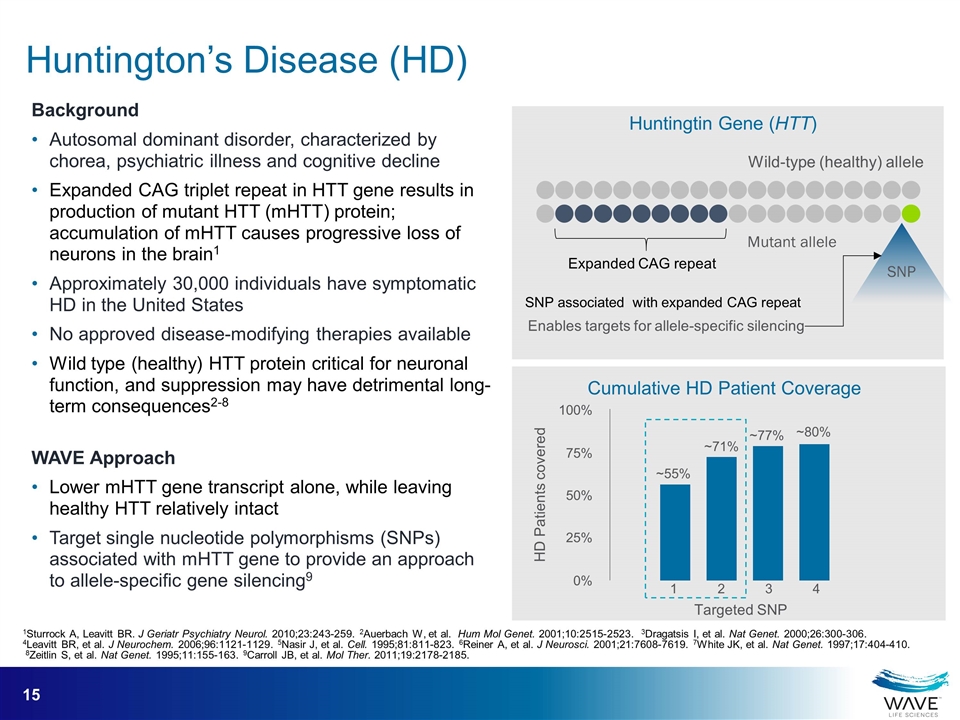
Huntingtin Gene (HTT) Expanded CAG repeat Wild-type (healthy) allele Mutant allele SNP associated with expanded CAG repeat SNP Enables targets for allele-specific silencing 100% 75% 25% 0% 1 2 3 4 Targeted SNP HD Patients covered Cumulative HD Patient Coverage ~80% 50% ~77% ~71% ~55% Background Autosomal dominant disorder, characterized by chorea, psychiatric illness and cognitive decline Expanded CAG triplet repeat in HTT gene results in production of mutant HTT (mHTT) protein; accumulation of mHTT causes progressive loss of neurons in the brain1 Approximately 30,000 individuals have symptomatic HD in the United States No approved disease-modifying therapies available Wild type (healthy) HTT protein critical for neuronal function, and suppression may have detrimental long-term consequences2-8 WAVE Approach Lower mHTT gene transcript alone, while leaving healthy HTT relatively intact Target single nucleotide polymorphisms (SNPs) associated with mHTT gene to provide an approach to allele-specific gene silencing9 Huntington’s Disease (HD) 1Sturrock A, Leavitt BR. J Geriatr Psychiatry Neurol. 2010;23:243-259. 2Auerbach W, et al. Hum Mol Genet. 2001;10:2515-2523. 3Dragatsis I, et al. Nat Genet. 2000;26:300-306. 4Leavitt BR, et al. J Neurochem. 2006;96:1121-1129. 5Nasir J, et al. Cell. 1995;81:811-823. 6Reiner A, et al. J Neurosci. 2001;21:7608-7619. 7White JK, et al. Nat Genet. 1997;17:404-410. 8Zeitlin S, et al. Nat Genet. 1995;11:155-163. 9Carroll JB, et al. Mol Ther. 2011;19:2178-2185.
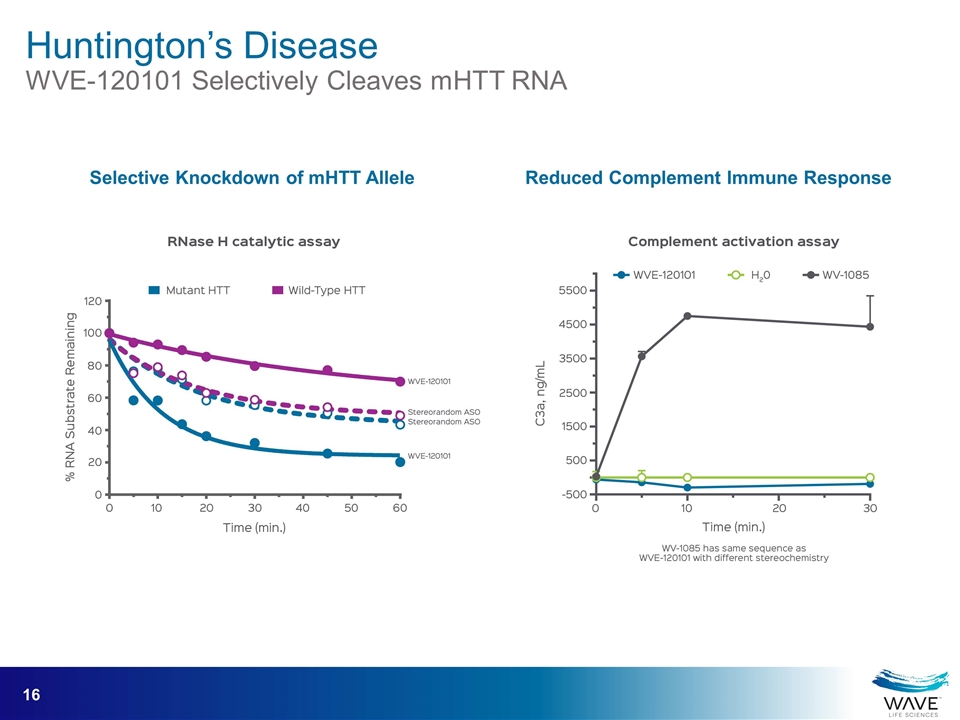
Huntington’s Disease WVE-120101 Selectively Cleaves mHTT RNA Reduced Complement Immune Response Selective Knockdown of mHTT Allele
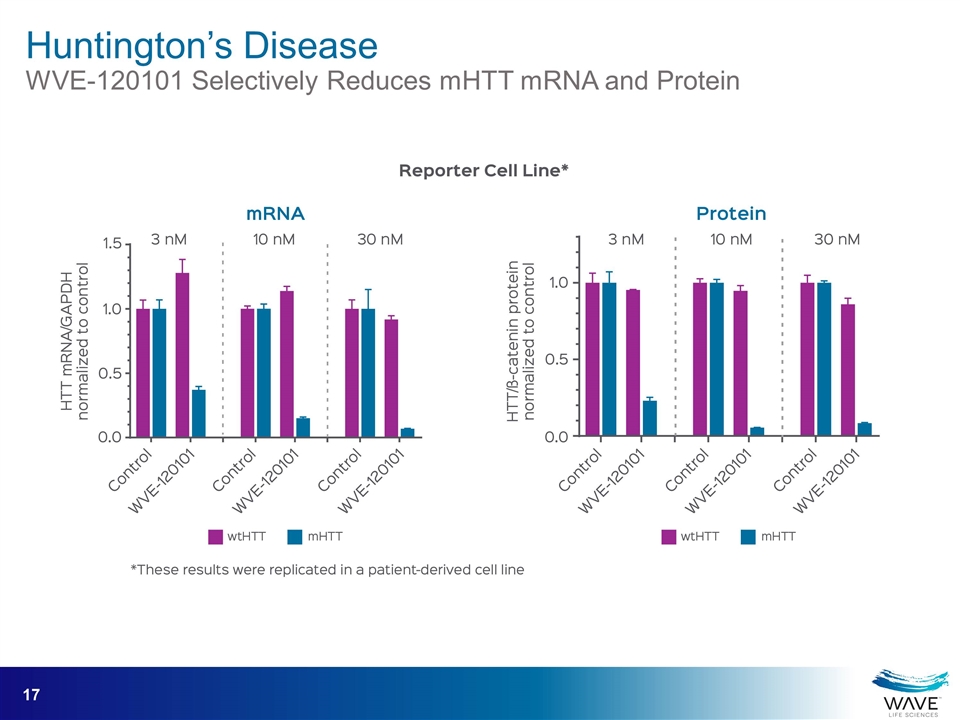
Huntington’s Disease WVE-120101 Selectively Reduces mHTT mRNA and Protein
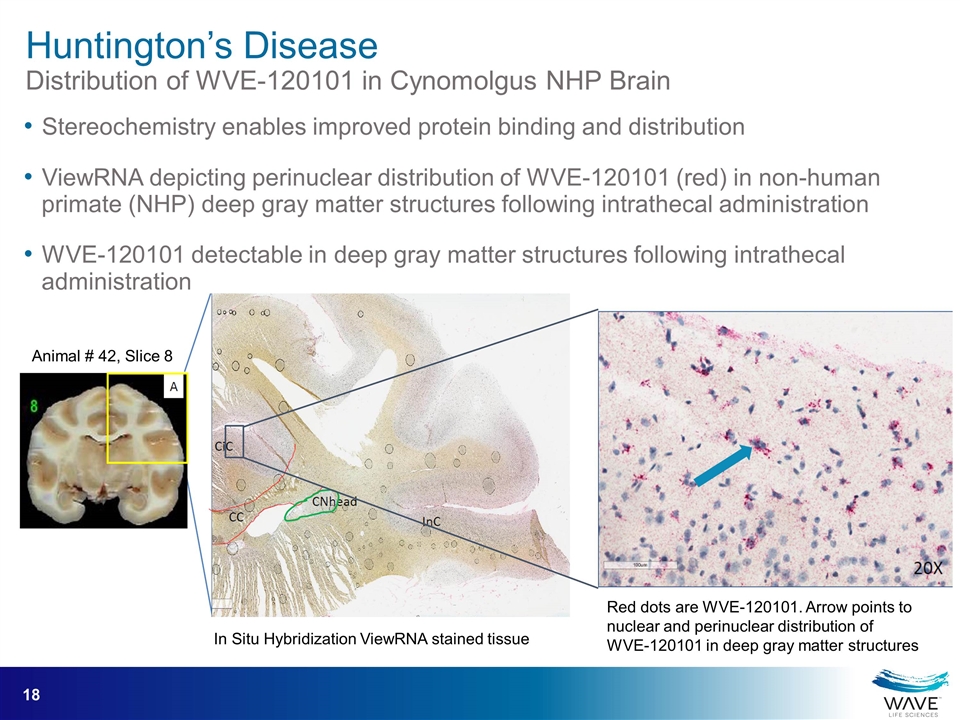
Stereochemistry enables improved protein binding and distribution ViewRNA depicting perinuclear distribution of WVE-120101 (red) in non-human primate (NHP) deep gray matter structures following intrathecal administration WVE-120101 detectable in deep gray matter structures following intrathecal administration Huntington’s Disease Distribution of WVE-120101 in Cynomolgus NHP Brain Animal # 42, Slice 8 Red dots are WVE-120101. Arrow points to nuclear and perinuclear distribution of WVE-120101 in deep gray matter structures In Situ Hybridization ViewRNA stained tissue
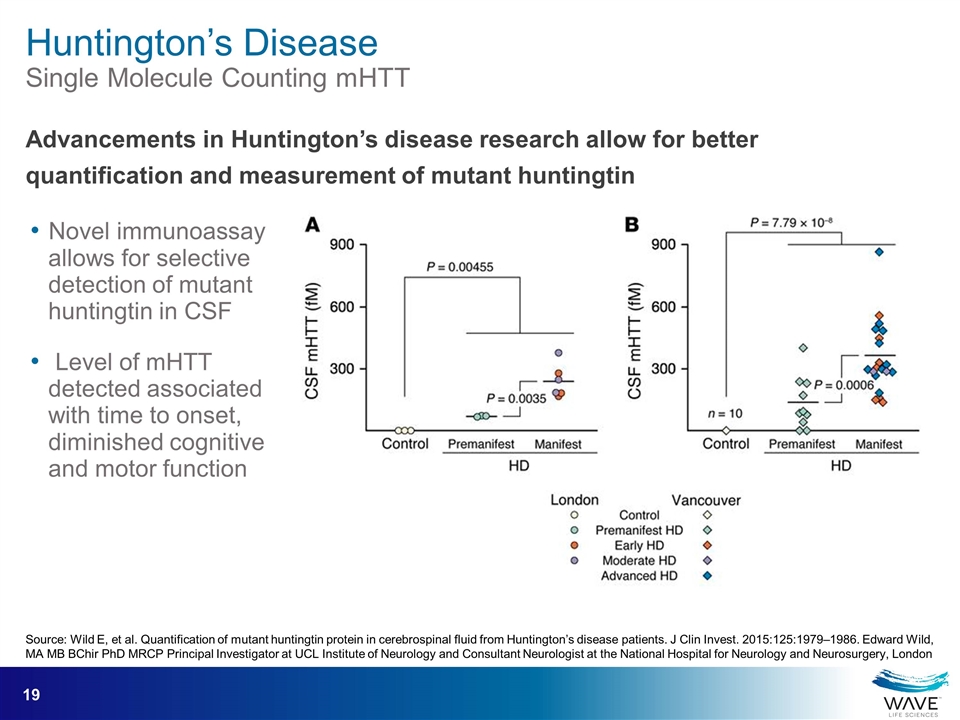
Source: Wild E, et al. Quantification of mutant huntingtin protein in cerebrospinal fluid from Huntington’s disease patients. J Clin Invest. 2015:125:1979–1986. Edward Wild, MA MB BChir PhD MRCP Principal Investigator at UCL Institute of Neurology and Consultant Neurologist at the National Hospital for Neurology and Neurosurgery, London Novel immunoassay allows for selective detection of mutant huntingtin in CSF Level of mHTT detected associated with time to onset, diminished cognitive and motor function Advancements in Huntington’s disease research allow for better quantification and measurement of mutant huntingtin Huntington’s Disease Single Molecule Counting mHTT

Refile IND for WVE-120101 and file IND for WVE-120102 in 1H 2017 with updated clinical protocols, accelerating time to multi-ascending dose (MAD) studies and potentially reducing time to proof-of-concept (POC) Updated plan informed by discussions with the FDA and new supportive data from multi-dose animal toxicology studies Updated plan includes moving straight to two simultaneous multi-ascending-dose (MAD) studies, potentially accelerating time to proof-of-concept Clinical Trial Applications in Europe on track for 1H 2017 First-in-patient dosing for both WVE-120101 (SNP-1) and WVE-120102 (SNP-2) trials expected mid-year 2017 20 Huntington’s Disease Clinical Development Update

1 Updated Clinical Trial Design for WVE-120101 and WVE-120102 Two parallel global placebo-controlled MAD trials targeting SNP-1 and SNP-2, respectively Primary Objective: Assess safety and tolerability of intrathecal doses in early manifest HD patients Additional Objectives: Exploratory pharmacokinetic, pharmacodynamic, clinical and MRI endpoints Patient SNP determination (SNP-1, SNP-2, other) at pre-screening visit Approximately 60 patients per trial Key inclusion criteria: Age ≥25 to ≤65, Stage I or Stage II Huntington’s disease Huntington’s Disease Clinical Development Update
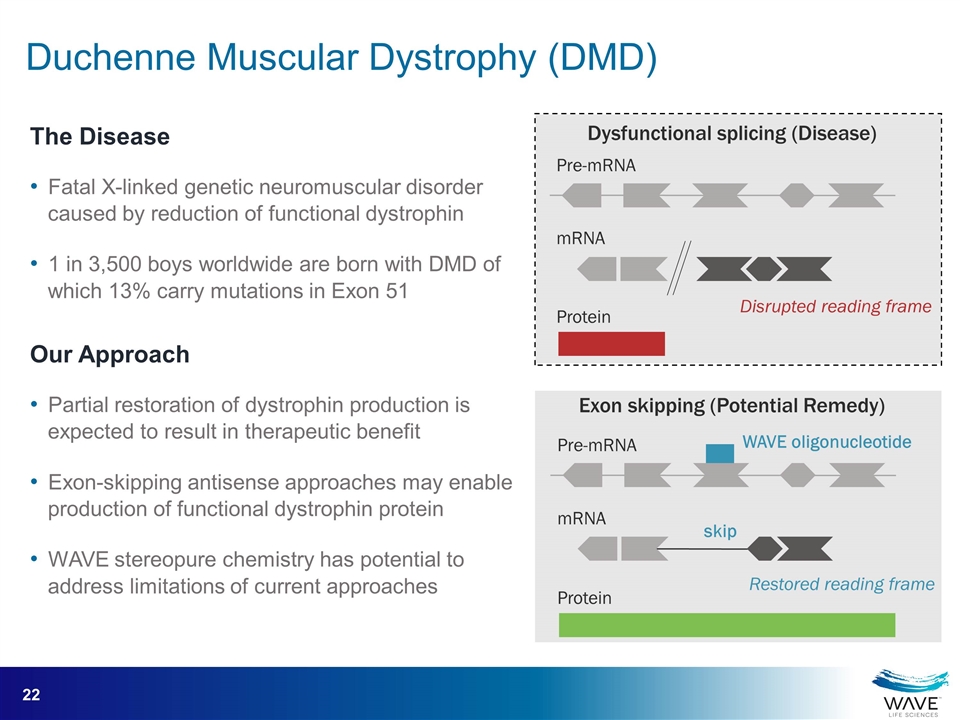
Pre-mRNA mRNA Protein Pre-mRNA mRNA Protein Exon skipping (Potential Remedy) WAVE oligonucleotide skip Disrupted reading frame Restored reading frame Dysfunctional splicing (Disease) The Disease Fatal X-linked genetic neuromuscular disorder caused by reduction of functional dystrophin 1 in 3,500 boys worldwide are born with DMD of which 13% carry mutations in Exon 51 Our Approach Partial restoration of dystrophin production is expected to result in therapeutic benefit Exon-skipping antisense approaches may enable production of functional dystrophin protein WAVE stereopure chemistry has potential to address limitations of current approaches Duchenne Muscular Dystrophy (DMD)
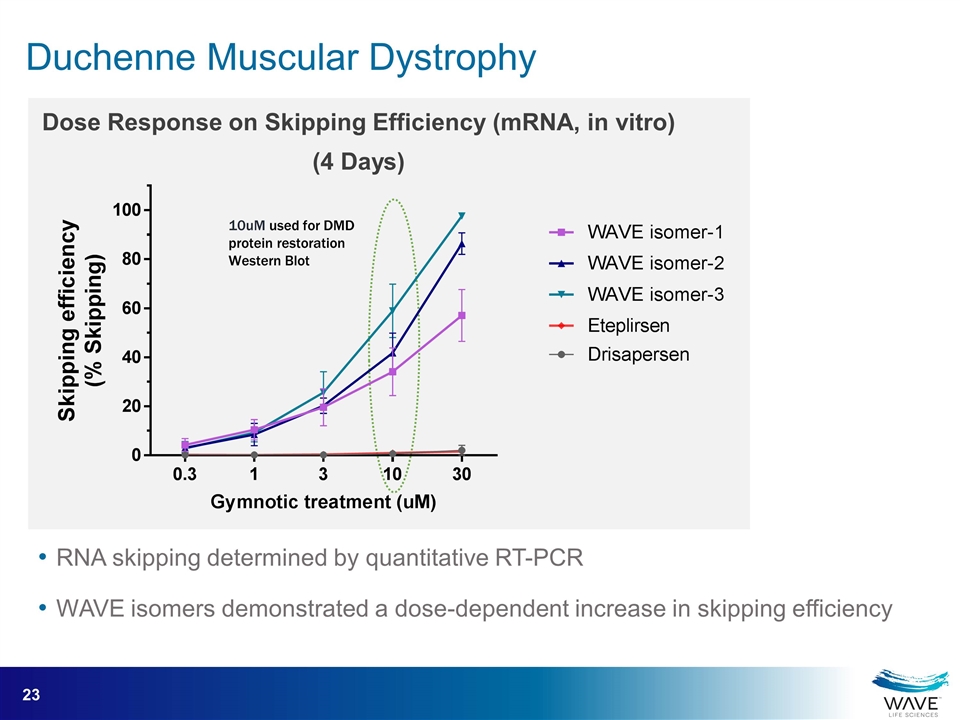
RNA skipping determined by quantitative RT-PCR WAVE isomers demonstrated a dose-dependent increase in skipping efficiency Dose Response on Skipping Efficiency (mRNA, in vitro) (4 Days) 10uM used for DMD protein restoration Western Blot Duchenne Muscular Dystrophy
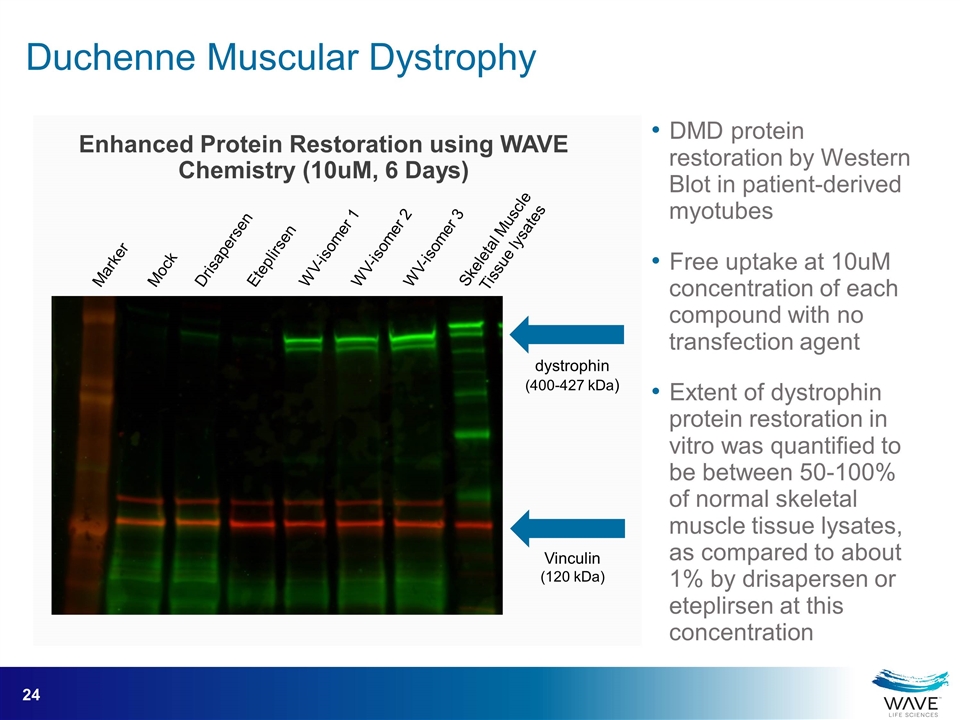
Enhanced Protein Restoration using WAVE Chemistry (10uM, 6 Days) Skeletal Muscle Tissue lysates Marker Mock Drisapersen Eteplirsen WV-isomer 1 WV-isomer 2 WV-isomer 3 dystrophin (400-427 kDa) Vinculin (120 kDa) DMD protein restoration by Western Blot in patient-derived myotubes Free uptake at 10uM concentration of each compound with no transfection agent Extent of dystrophin protein restoration in vitro was quantified to be between 50-100% of normal skeletal muscle tissue lysates, as compared to about 1% by drisapersen or eteplirsen at this concentration Duchenne Muscular Dystrophy
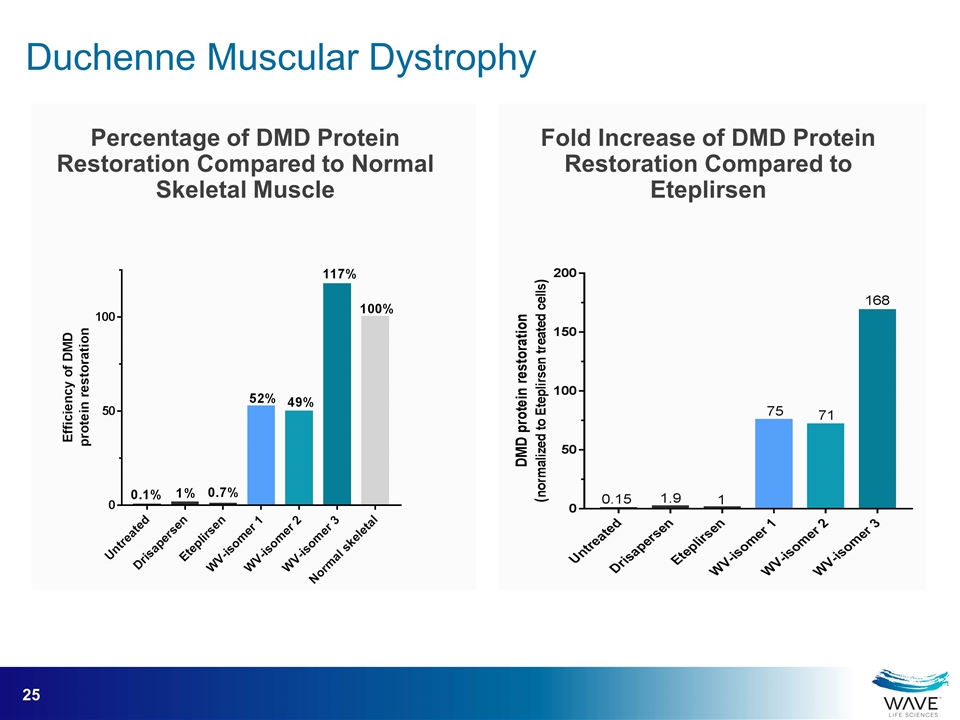
Fold Increase of DMD Protein Restoration Compared to Eteplirsen Percentage of DMD Protein Restoration Compared to Normal Skeletal Muscle Duchenne Muscular Dystrophy

Candidate selected for DMD Exon 51, WVE-210201 (WV-isomer 1) Clinical trials on track to initiate 2H 2017 Protocol development in collaboration with DMD community Trials to include ambulatory and non-ambulatory patients Patients previously treated with other exon skipping therapies will not be excluded Measurement of dystrophin via standardized Western Blot Initiation of GMP manufacturing Duchenne Muscular Dystrophy Clinical Development Update

27 GMP manufacturing facility leased in Lexington, MA Increases control and visibility of our manufacturing supply chain, with potential commercial-scale capabilities by 2019 90,000 square foot facility to support future growth Supplements existing Cambridge headquarters and facilities Facility expected to be occupied 2Q 2017 WAVE Manufacturing Capabilities

2017 Upcoming Catalysts Initiating Three Clinical Programs in 2017 Initiate two clinical trials in Huntington’s disease mid-2017 Potential to be first two allele-specific disease-modifying therapies Initiate clinical trial in DMD 2H 2017 First stereopure oligonucleotide targeting Exon 51 with potential to be best-in-class Nominate three additional proprietary therapeutic candidates to progress to clinic Initiate in-house GMP production of stereopure oligonucleotide candidates 28
